Team:EPF-Lausanne/Protocols/TetR
From 2011.igem.org
(→PCR Cycle (30 cycles)) |
|||
| (9 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{:Team:EPF-Lausanne/Templates/ | + | {{:Team:EPF-Lausanne/Templates/ProtocolHeader|title=Linear template- TetR}} |
Two step extension PCR | Two step extension PCR | ||
| Line 26: | Line 26: | ||
| - | + | :<b>1.</b> 98°C for 30s | |
| - | + | :<b>2.</b> 98°C for 7.5 seconds | |
| - | + | :<b>3.</b> 58°C for 20 sec (this depends on the primers, | |
| - | therefore look up the annealing temperature of the primers used | + | therefore look up the annealing temperature of the primers used |
| - | + | :<b>4.</b> 72°C for 15 sec; you have to calculate the time required. | |
This polymerase has an effciency of 15-30s/kb.(calculate for the longest fragment) | This polymerase has an effciency of 15-30s/kb.(calculate for the longest fragment) | ||
| - | + | :<b>5.</b> 72°C for 5 min | |
| - | + | :<b>6.</b> 4°C "forever" | |
*Save the file in the MAIN folder→ OK→ SAVE→RUN→Select the block you use→ RUN→ Change sample volume to 50µl, LidT should be 105°C and the box just below ("hotlid") should ALWAYS be checked | *Save the file in the MAIN folder→ OK→ SAVE→RUN→Select the block you use→ RUN→ Change sample volume to 50µl, LidT should be 105°C and the box just below ("hotlid") should ALWAYS be checked | ||
*Press STATUS to check the progress | *Press STATUS to check the progress | ||
| - | |||
== <b>Extension PCR</b> == | == <b>Extension PCR</b> == | ||
== Materials == | == Materials == | ||
| - | + | * 10µl 5x iProof HF buffer | |
| - | + | * 0.5 µl of each extension primer@500nM | |
| - | + | * 1 µl dNTP mix of 10mM | |
| - | + | * 0.5 µl High Fidelity Polymerase | |
| - | + | * 1 µl of gene specific PCR product from previous step | |
| - | + | * 0.5 µl Polymerase | |
| - | + | * 36.5 µl H2O | |
Extension Primers: | Extension Primers: | ||
| Line 58: | Line 57: | ||
== PCR Cycle (10 cycles) == | == PCR Cycle (10 cycles) == | ||
| - | + | :<b>1.</b> 98°C for 30s | |
| - | + | :<b>2.</b> 98°C for 7.5 seconds | |
| - | + | :<b>3.</b> 58°C for 20 sec | |
| - | + | :<b>4.</b> 72°C for 15 sec; | |
| - | + | :<b>5.</b> 72°C for 5 min | |
| - | + | :<b>6.</b> 4°C "forever" | |
| - | + | ||
== Materials == | == Materials == | ||
| - | + | * 10µl 5x iProof HF buffer | |
| - | + | * 0.5 µl of each final primer @50µM/1µl of primer mix@1µM | |
| - | + | * 1 µl dNTP mix of 10mM | |
| - | + | * 0.5 µl High Fidelity Polymerase | |
| - | + | * 1 µl of previous PCR product(extension PCR) | |
| - | + | * 0.5 µl Polymerase | |
| - | + | * 36.5 µl H2O | |
Final primers | Final primers | ||
| Line 84: | Line 82: | ||
== PCR Cycle (30 cycles) == | == PCR Cycle (30 cycles) == | ||
| - | + | :<b>1.</b> 98°C for 30s | |
| - | + | :<b>2.</b> 98°C for 7.5 seconds | |
| - | + | :<b>3.</b> 53°C for 20 sec | |
| - | + | :<b>4.</b> 72°C for 15 sec; | |
| - | + | :<b>5.</b> 72°C for 5 min | |
| - | + | :<b>6.</b> 4°C "forever" | |
{{:Team:EPF-Lausanne/Templates/Footer}} | {{:Team:EPF-Lausanne/Templates/Footer}} | ||
Latest revision as of 08:48, 15 July 2011
Linear template- TetR
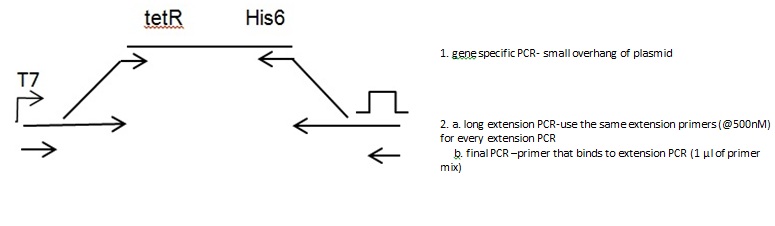
Back to protocols.Two step extension PCR
Contents |
Gene PCR
Materials
- 10µl 5x iProof HF buffer
- 0.5 µl of each primer
- 1 µl dNTP mix of 10mM
- 0.5 µl High Fidelity Polymerase
- 1 µl DNA template
- 0.5 µl Polymerase
- 36.5 µl H2O
Primers: TetR Forward: CTC GAG AAT TCG CCA CCA TGT CCA GAT TAG ATA AAA GTA AAG ; Tm=62.6ºC
TetR-His6 Reverse: GTA GCA GCC TGA GTC GTT ATT AAT GAT GAT GAT GAT GAT GAG AAC CCC CAG ACC CAC TTT CAC ATT TAA G ; Tm=68.5 ºC
PCR Cycle (30 cycles)
- 1. 98°C for 30s
- 2. 98°C for 7.5 seconds
- 3. 58°C for 20 sec (this depends on the primers,
therefore look up the annealing temperature of the primers used
- 4. 72°C for 15 sec; you have to calculate the time required.
This polymerase has an effciency of 15-30s/kb.(calculate for the longest fragment)
- 5. 72°C for 5 min
- 6. 4°C "forever"
- Save the file in the MAIN folder→ OK→ SAVE→RUN→Select the block you use→ RUN→ Change sample volume to 50µl, LidT should be 105°C and the box just below ("hotlid") should ALWAYS be checked
- Press STATUS to check the progress
Extension PCR
Materials
- 10µl 5x iProof HF buffer
- 0.5 µl of each extension primer@500nM
- 1 µl dNTP mix of 10mM
- 0.5 µl High Fidelity Polymerase
- 1 µl of gene specific PCR product from previous step
- 0.5 µl Polymerase
- 36.5 µl H2O
Extension Primers:
Forward: GAT CTT AAG GCT AGA GTA CTA ATA CGA CTC ACT ATA GGG AAT ACA AGC TAC TTG TTC TTT TTG CAC TCG AGA ATT CGC CAC C Tm=68.4 ºC
Reverse: CAA AAA ACC CCT CAA GAC CCG TTT AGA GGC CCC AAG GGG TTA TGC TAG TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT GTA GCA GCC TGA GTC G; Tm=70.4ºC
PCR Cycle (10 cycles)
- 1. 98°C for 30s
- 2. 98°C for 7.5 seconds
- 3. 58°C for 20 sec
- 4. 72°C for 15 sec;
- 5. 72°C for 5 min
- 6. 4°C "forever"
Materials
- 10µl 5x iProof HF buffer
- 0.5 µl of each final primer @50µM/1µl of primer mix@1µM
- 1 µl dNTP mix of 10mM
- 0.5 µl High Fidelity Polymerase
- 1 µl of previous PCR product(extension PCR)
- 0.5 µl Polymerase
- 36.5 µl H2O
Final primers
Forward: GAT CTT AAG GCT AGA GTA C; Tm=46.0ºC
Reverse: CAA AAA ACC CCT CAA GAC; Tm=48.6ºC
PCR Cycle (30 cycles)
- 1. 98°C for 30s
- 2. 98°C for 7.5 seconds
- 3. 53°C for 20 sec
- 4. 72°C for 15 sec;
- 5. 72°C for 5 min
- 6. 4°C "forever"
 "
"