Team:UTP-Panama/After Regional Week 2
From 2011.igem.org
(→October 25, 2011) |
|||
| (25 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:UTP-Panama/utp_calendar_template}} | {{Team:UTP-Panama/utp_calendar_template}} | ||
== Octuber 24 to 28== | == Octuber 24 to 28== | ||
| - | . | + | ===October 24, 2011=== |
| + | '''Morning'''<br> | ||
| + | This day there was made an electrophoresis of digestion products which became on September 22, 2011 (the upstream and downstream). Following the protocol for electrophoresis:<br> | ||
| + | 1. Preparation of 1% agarose. <br> | ||
| + | 2. Moisten the chamber walls. <br> | ||
| + | 3. In a 70 mL Erlenmeyer flask add the agar and put a minute in the microwave, then take it off shake it and put it in the microwave 20 seconds. <br> | ||
| + | 4. Ethidium bromide placed in the microwave and cooled with tap water to a temperature between 60 ° C and 70 º C. | ||
| + | 5. Placed between 2.5 to 5 micolitros Ethidium bromide. We mix before pouring it into the camera. <br> | ||
| + | 6. Once the gel solidified cover it with TAE (1X). (A) <br> | ||
| + | 7. 35μL were added ultra-filtered water plasmids were 65.5 ° C.<br> | ||
| + | 8. Droplets were eliminated on the walls with the sping-down applied to the tubes. <br> | ||
| + | 9. We put the tubes in the microwave 5 minutes. <br> | ||
| + | 10. Add loading buffer which was frozen thawed. How?<br> | ||
| + | • Put dots of paper loading buffer. <br> | ||
| + | • Of the 35 uL of plasmid take 10 mL (B).<br> | ||
| + | • Re mix in the dots and then the wells. <br> | ||
| + | 11. A and B meet. Plasmid preparation and gel electrophoresis.<br> | ||
| + | 12. The plasmids with the loading buffer in the wells <br> | ||
| + | Note: It is important to place the (-) side of the DNA <br> | ||
| + | 13. We ran the gel for 2 hours.<br> | ||
| + | Our Upstream is BBa_K410000 and the Downstream is BBa_K381001<br> | ||
| + | <br> | ||
| + | '''Afternoon''' | ||
| + | At this time a purification protocol was required but in the gel electrophoresis previously made was not enough genetic material to recover so do not purified at this time. <br> | ||
| + | To remember our goal with these protocols a flow chart was made and is presented below.<br> | ||
| + | <br> | ||
| + | |||
| + | <div align="center"> | ||
| + | [[File:Diagramaflujowet.jpg]] | ||
| + | |||
| + | <div align="justify"> | ||
| + | |||
| + | ===October 25, 2011=== | ||
| + | |||
| + | We ran an electrophoresis before we started the purification. The bands we needed were the ones with the weight mentioned. | ||
| + | |||
| + | |||
| + | Information necessary for the purification:<br> | ||
| + | BBa_K381001: 986bp<br> | ||
| + | BBa_K410000: 1517bp<br> | ||
| + | pSB1C3: 2070bp<br> | ||
| + | |||
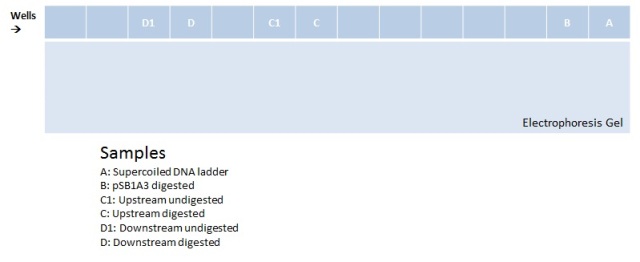
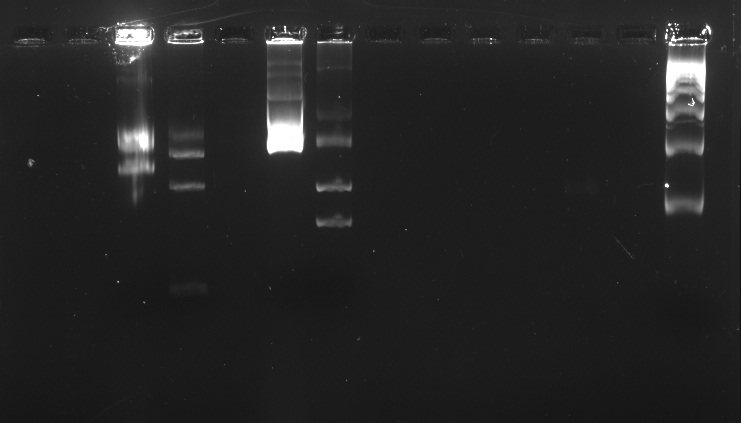
| + | Electrophoresis with BBa_K410000, BBa_K381001, and pSB1A3 digested and undigested<br> | ||
| + | <br> | ||
| + | |||
| + | [[File:Gelexplanutp.jpg]] | ||
| + | [[File:Electrof25.jpg]] | ||
| + | |||
| + | In the well labeled as "B" of the picture there is nothing observed because the amount of sample put was very little. | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
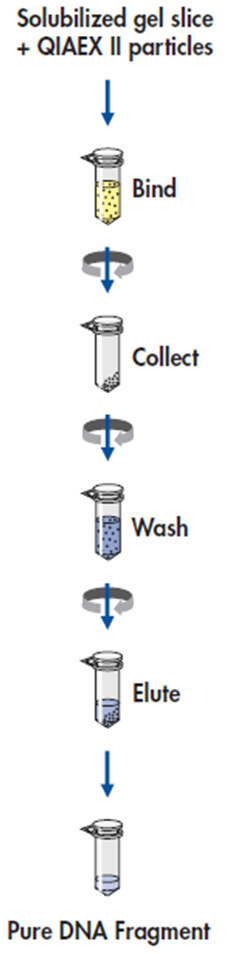
| + | '''Purification'''<br> | ||
| + | Protocol: Extraction of 40 bp to 50 kb DNA fragments from Agarose Gels<br> | ||
| + | Procedure<br> | ||
| + | 1. DNA band was excised from the agarose gel with a clean, sharp scalpel. It was used a 2ml microfuge tube.<br> | ||
| + | 2. Gel slice was weighed in a colorless tube. Then added 3 volumes of Buffer QX1 to 1 volume of gel for DNA fragments 100 bp – 4 kb. For us, added 600 μl of Buffer QX1 to 200 mg of gel.<br> | ||
| + | 3. Re-suspended QIAEX II by vortexing for 30 s. Added 500μl QIAEX II to the sample.<br> | ||
| + | 4. Incubated at 50°C for 10 min to solubilize the agarose and to bind the DNA. Mixed by vortexing every 2min to keep QIAEX II in suspension. Checked that the color of the mixture is yellow.<br> | ||
| + | 5. Sample was centrifuged for 1min at 10000rpm and carefully removed supernatant with a pipet.<br> | ||
| + | 6. Pellet was washed with 500μl of Buffer QX1. Discarded supernatant. Then it was centrifuged.<br> | ||
| + | 7. Pellet was washed twice with 500μl of Buffer PE. Discarded supernatant. Then it was centrifuged.<br> | ||
| + | 8. Air-dried pellet for 10–15 min or until the pellet becomes white.<br> | ||
| + | 9. DNA was eluted, added 20μl of H2O and re-suspended the pellet by vortexing. This was incubated at room temperature for 5 min.<br> | ||
| + | 10. It was centrifuged for 1min, then pipet the supernatant into a clean tube.<br> | ||
| + | <br> | ||
| + | <div align="center">PURIFICATION PROTOCOL<br> | ||
| + | [[File:Purifprot.png]]</div> | ||
| + | Source: | ||
| + | QIAEX II® Handbook. For DNA extraction from agarose and polyacrylamide gels and for desalting and concentrating DNA from solutions. October 2008. QIAGEN Sample and Assay Technologies. | ||
Latest revision as of 03:24, 29 October 2011
|
Home |
Week 1 | Week 2 | Week 3 | Week 4 | Week 5 | Week 6 | Week 7 | Week 8 | Week 9 | Week 10 | Week 11 | Week 12 | Week 13 | Week 14 | Week 15 | Week 16 | Week 17 | After Regional Week 1 | After Regional Week 2 | Octuber 24 to 28October 24, 2011Morning October 25, 2011We ran an electrophoresis before we started the purification. The bands we needed were the ones with the weight mentioned.
Electrophoresis with BBa_K410000, BBa_K381001, and pSB1A3 digested and undigested In the well labeled as "B" of the picture there is nothing observed because the amount of sample put was very little.
Purification Source: QIAEX II® Handbook. For DNA extraction from agarose and polyacrylamide gels and for desalting and concentrating DNA from solutions. October 2008. QIAGEN Sample and Assay Technologies. |
 "
"