Team:EPF-Lausanne/Notebook/October2011
From 2011.igem.org
(→Tuesday, October 25 2011) |
|||
| (11 intermediate revisions not shown) | |||
| Line 37: | Line 37: | ||
For the K1 backbone, the recipe was slightly different due to the higher concentration of blunted DNA we wanted to achieve: | For the K1 backbone, the recipe was slightly different due to the higher concentration of blunted DNA we wanted to achieve: | ||
| - | + | * H20 - 4.4 uL | |
| + | * NEBuffer2 (10X, found in enzyme ice box) - 4 uL (needs to be at a final concentration of 1X) | ||
| + | * dNTP (10 mM) - 0.4 (needs to be at a final concentration of 100 uM) | ||
| + | * template - 30 uL (for 1.5 ug of DNA, assuming the T7 + Lysis was at a concentration of 100 ng/uL) | ||
| + | * T4 Polymerase - 1.2 uL (it comes at a "concentration" of 3000 units/mL and we want 1.5 units) | ||
| + | for a total of 40 uL. | ||
| + | |||
| + | == Sunday, October 23 2011 == | ||
| + | |||
| + | Vincent ran a colony PCR on the 1 and 13 T7 promoter + Lysis + psB3K1 constructs, since they were the ones that yielded the highest colony count (compared to the negative control). He chose ten colonies from each plate. The protocol was as followed: | ||
| + | |||
| + | |||
| + | * TPB 2.5 uL | ||
| + | * EPFL Taq 0.25 uL | ||
| + | * primer FW (10 uM) 0.5 uL (1289_F, originally 100 uM) | ||
| + | * primer RV (10 uM) 0.5 uL (pSB-Pcon-TetR-r, originally 20 uM) | ||
| + | * template 2 uL of DNA (boiled) | ||
| + | * dNTP 0.5 uL | ||
| + | * dH20 18.75 uL | ||
| + | _________________________ | ||
| + | 25 uL total | ||
| + | |||
| + | The cycle (now called EPFLCOP for EPFL Colony PCR) is as follows: | ||
| + | |||
| + | * 94 °C, 2 minutes | ||
| + | 30 times: | ||
| + | * 94 °C, 15 sec. | ||
| + | * 58 °C, 15 sec. | ||
| + | * 68 °C, 1 minute (1 min / kb, and we expect 629 bp) | ||
| + | End of 30 | ||
| + | * 68 °C, 5 min | ||
| + | * 10 °C, infinitely | ||
| + | |||
| + | |||
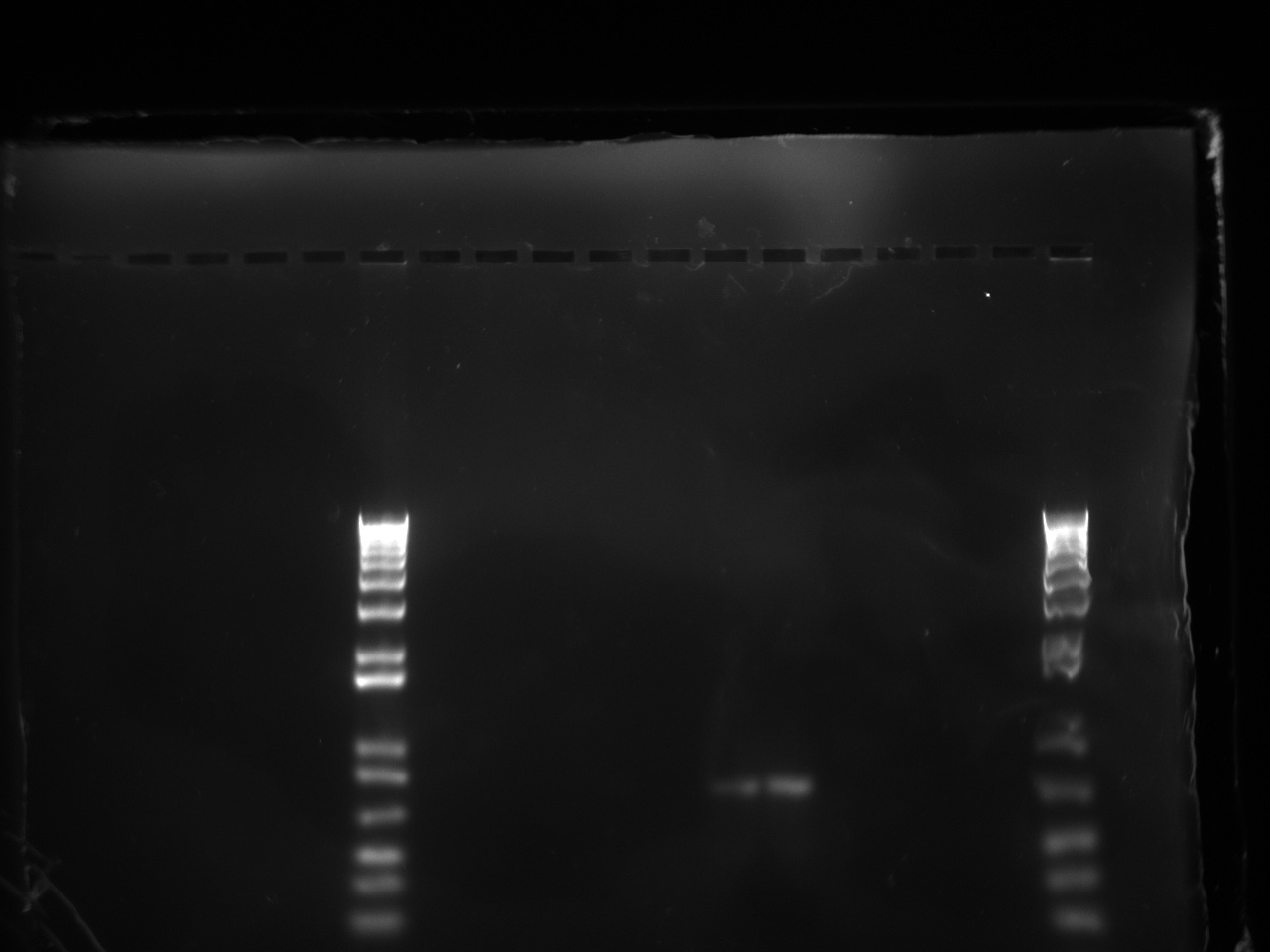
| + | The gel showed a strong signal at 630 bp for two colonies of T7-1 but none for T7-13. Unfortunately, this is the same promoter as the one amplified by Alina two months ago, so we can't use it to run an interesting experiment. | ||
| + | |||
| + | [[File:Trashed t7promgel.jpg|500px]] | ||
| + | |||
| + | == Monday, October 24 2011 == | ||
| + | |||
| + | Vincent mini-prepped the liquid culture from the successful T71-Lysis-K1 plasimd identified by PCR the previous day. He then transformed that DNA into BL21 cells and plated the transformant. | ||
| + | |||
| + | Since the primers used on Sunday would miss half of the inserts (the blunt-end strategy implies that using primers on the insert and on the vector will miss half of the potentially good plasmids), we chose to switch to different primers: the 816_R and 30_F which amplify on the lysis cassette. | ||
| + | |||
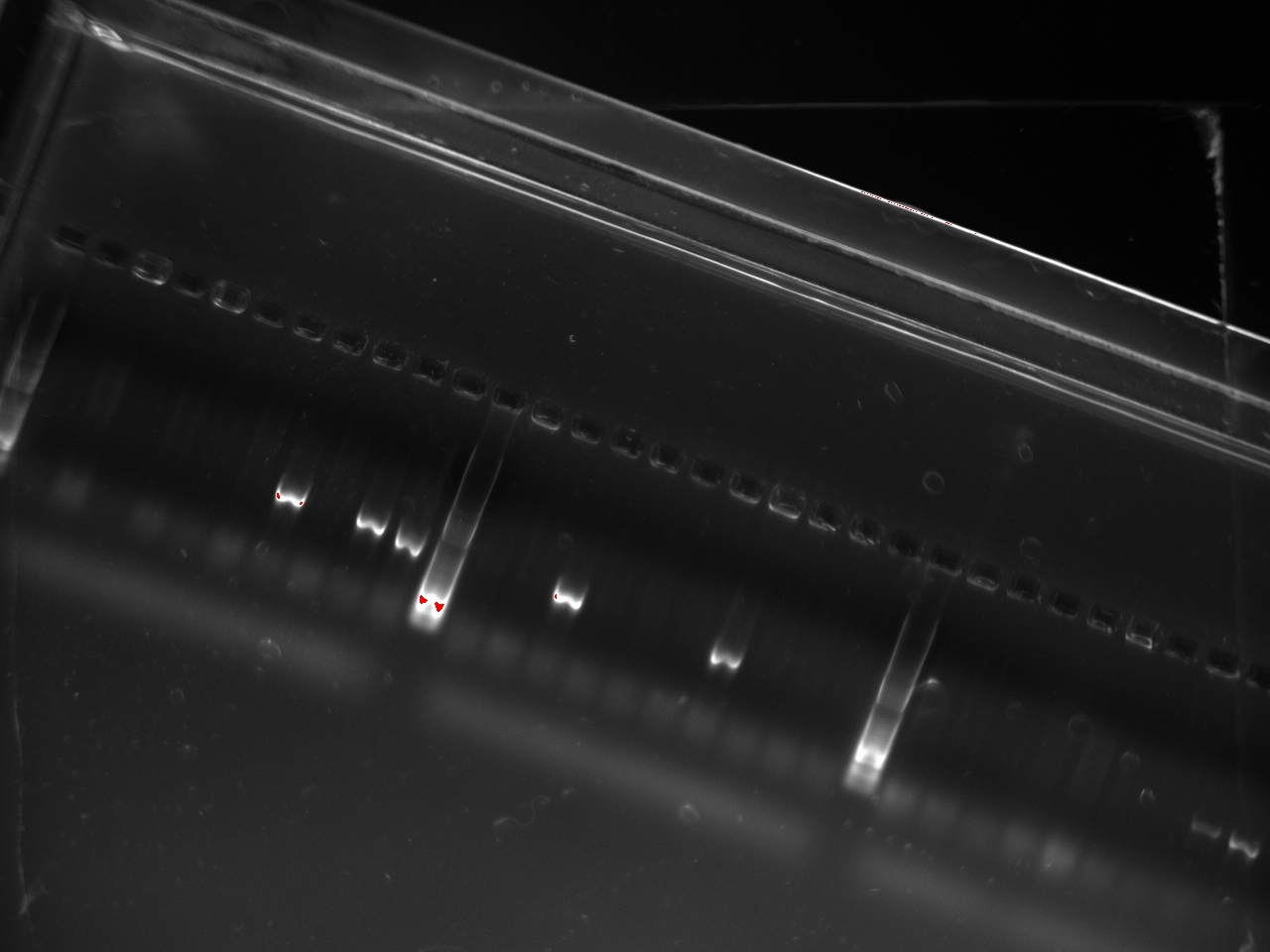
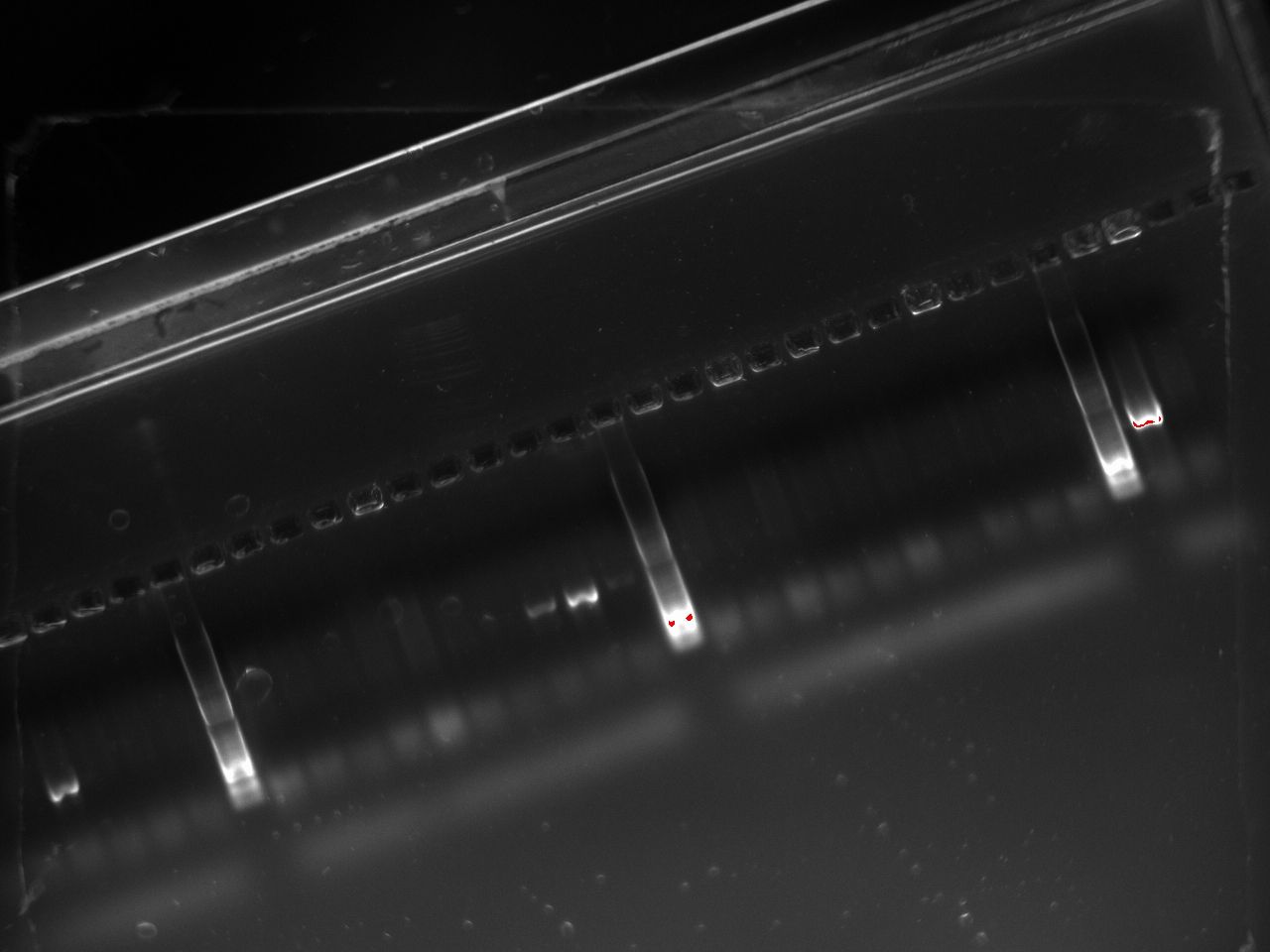
| + | With these primers, Vincent ran a colony PCR on 10 colonies per plate for the 2, 3, 4, and 6 plates (2,3,4, and 6 refers to the T7 promoter type). He used the same protocol as on Sunday. The resulting gel shows that 3 out of the 4 T7 variants show an amplification on some colony: | ||
| + | |||
| + | [[File:2011-10-24-t7-ligation-colPCR-1.jpg|400px]] | ||
| + | |||
| + | |||
| + | [[File:2011-10-24-t7-ligation-colPCR-2.jpg|400px]] | ||
| + | |||
| + | == Tuesday, October 25 2011 == | ||
| + | |||
| + | Nadine and Douglas came in and mini-prepped liquid cultures prepared the previous day by Nadine. | ||
| + | |||
| + | Vincent transformed the resulting DNA into BL21 cells. | ||
| + | |||
| + | |||
| + | == Wednesday, October 25 2011 == | ||
| + | |||
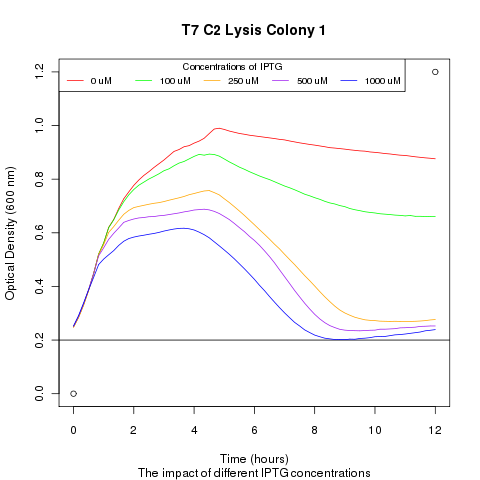
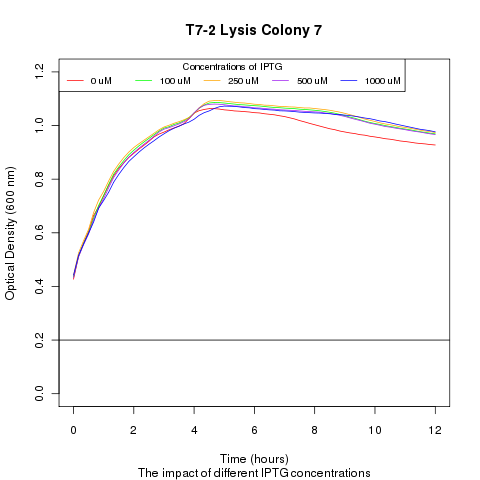
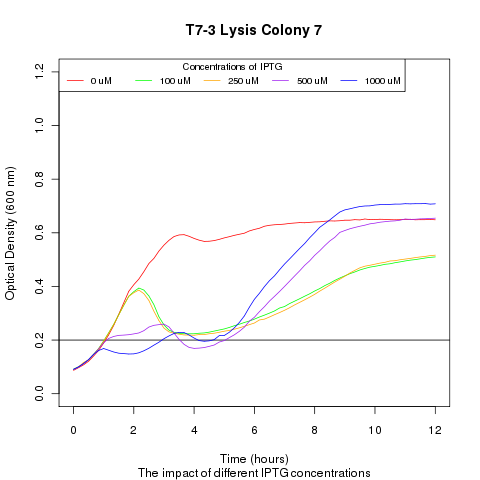
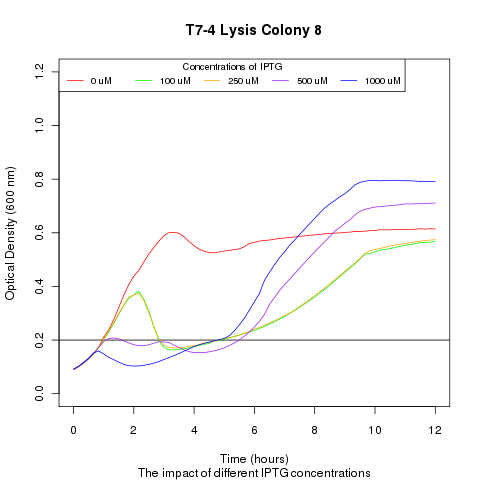
| + | Late on Tuesday, Henrike noticed that the colonies had grown sufficiently to make liquid cultures for a platereader. Nadine ran the platereader on Wednesday with the following results: | ||
| + | |||
| + | [[File:t7_c2_lysis_col1.png|400px]] | ||
| + | [[File:t7_2_lysis_col7.png|400px]] | ||
| + | [[File:t7_3_lysis_col3.png|400px]] | ||
| + | [[File:t7_4_lysis_col8.png|400px]] | ||
{{:Team:EPF-Lausanne/Templates/Footer}} | {{:Team:EPF-Lausanne/Templates/Footer}} | ||
Latest revision as of 06:54, 28 October 2011
Notebook: October 2011
Contents |
Friday, October 7 2011
Matt did gene-specific PCR -- touchdown-style -- on the lysis cassette. The gel results look fantastic. Pictures coming soon. Next up, adding the T7 promoters (all twelve of them) upstream of lysis.
Monday, October 10 2011
Over the weekend, Matt and Vincent figured out how the PCRs ought to work out to get the different T7 promoters upstream of the lysis cassette, which had been successfully amplified earlier last week. A helpful website indicated what the right melting temperatures should be. From there, Matt kindly took the initiative of making all 12 T7-lysis combinations via PCR. He then ran a gel and took a bunch of sweet pictures, and the results look great: no additional PCR products and some very clean bands of the right size.
Wednesday, October 12 2011
Vincent ran a PCR to amplify pSB3K1, which we plan on using as a backbone for the T7 + Lysis inserts. He used Expand High Fidelity Polymerase and its associated protocol for an expected band of 2774 bp. The gel to check it will be run on Thursday.
Thursday, October 13 2011
The gel for the K1 PCR amplification was run, and the results look good: a nice thick band right below the 3000 bp mark. Now we have to put the T7 + Lysis together with the backbone, and for that there will have to be some blunt-ends with phosphorylation before we can ligate the two pieces together. Stay tuned.
Sunday, October 16 2011
Vincent came in to work on the blunt-end cloning for the inserts (T7 promoters + Lysis) and the psB3K1 vector. The idea is to use the T4 DNA polymerase, which has a 3' to 5' exonuclease, to fill-in and chew away the ends till we have blunt ends. I added the protocol to the appropriate protocol section. After running through this protocol, the T7 inserts had an average concentration of 40 ng/uL while the backbone vector K1 had a concentration of 90 ng/uL (to within Nanodrop measurement error). Those concentrations are high enough to allow us to move on to the next step which is phosphorylating the T7 inserts (not the backbone!) for ligation.
The phosphorylation step is needed because the ligation (via ligase enzyme) requires that one of the 5' termini have a phosphate residue that will induce phosphodiester bond. If both the insert and the backbone have the phosphate residue, then the backbone may bind to itself, leaving the insert out -- which would defeat the purpose of ligation.
The specifics of the blunt-end protocol were as follows:
For the T7 + Lysis inserts,
- H20 - 11.25 uL
- NEBuffer2 (10X, found in enzyme ice box) - 3 uL (needs to be at a final concentration of 1X)
- dNTP (10 mM) - 0.3 (needs to be at a final concentration of 100 uM)
- template - 15 uL (for 1.5 ug of DNA, assuming the T7 + Lysis was at a concentration of 100 ng/uL)
- T4 Polymerase - 0.45 uL (it comes at a "concentration" of 3000 units/mL and we want 1.5 units)
I made a master mix of everything except the template DNA, which then went into separate PCR tubes. The total is a 30 uL reaction. The tubes were placed in the Bio-Rad machine were we setup a protocol (called BLUNTEND) that keeps the tubes at a constant 12 C for 15 minutes. If you want, you can add EDTA to a final concentration of 10 mM (as advised by New England Biolabs) and heat activate the samples at 75 C for 20 minutes. We decided that it was not necessary in our case.
For the K1 backbone, the recipe was slightly different due to the higher concentration of blunted DNA we wanted to achieve:
- H20 - 4.4 uL
- NEBuffer2 (10X, found in enzyme ice box) - 4 uL (needs to be at a final concentration of 1X)
- dNTP (10 mM) - 0.4 (needs to be at a final concentration of 100 uM)
- template - 30 uL (for 1.5 ug of DNA, assuming the T7 + Lysis was at a concentration of 100 ng/uL)
- T4 Polymerase - 1.2 uL (it comes at a "concentration" of 3000 units/mL and we want 1.5 units)
for a total of 40 uL.
Sunday, October 23 2011
Vincent ran a colony PCR on the 1 and 13 T7 promoter + Lysis + psB3K1 constructs, since they were the ones that yielded the highest colony count (compared to the negative control). He chose ten colonies from each plate. The protocol was as followed:
- TPB 2.5 uL
- EPFL Taq 0.25 uL
- primer FW (10 uM) 0.5 uL (1289_F, originally 100 uM)
- primer RV (10 uM) 0.5 uL (pSB-Pcon-TetR-r, originally 20 uM)
- template 2 uL of DNA (boiled)
- dNTP 0.5 uL
- dH20 18.75 uL
_________________________ 25 uL total
The cycle (now called EPFLCOP for EPFL Colony PCR) is as follows:
- 94 °C, 2 minutes
30 times:
- 94 °C, 15 sec.
- 58 °C, 15 sec.
- 68 °C, 1 minute (1 min / kb, and we expect 629 bp)
End of 30
- 68 °C, 5 min
- 10 °C, infinitely
The gel showed a strong signal at 630 bp for two colonies of T7-1 but none for T7-13. Unfortunately, this is the same promoter as the one amplified by Alina two months ago, so we can't use it to run an interesting experiment.
Monday, October 24 2011
Vincent mini-prepped the liquid culture from the successful T71-Lysis-K1 plasimd identified by PCR the previous day. He then transformed that DNA into BL21 cells and plated the transformant.
Since the primers used on Sunday would miss half of the inserts (the blunt-end strategy implies that using primers on the insert and on the vector will miss half of the potentially good plasmids), we chose to switch to different primers: the 816_R and 30_F which amplify on the lysis cassette.
With these primers, Vincent ran a colony PCR on 10 colonies per plate for the 2, 3, 4, and 6 plates (2,3,4, and 6 refers to the T7 promoter type). He used the same protocol as on Sunday. The resulting gel shows that 3 out of the 4 T7 variants show an amplification on some colony:
Tuesday, October 25 2011
Nadine and Douglas came in and mini-prepped liquid cultures prepared the previous day by Nadine.
Vincent transformed the resulting DNA into BL21 cells.
Wednesday, October 25 2011
Late on Tuesday, Henrike noticed that the colonies had grown sufficiently to make liquid cultures for a platereader. Nadine ran the platereader on Wednesday with the following results:
 "
"