Team:Arizona State/Results/Data
From 2011.igem.org
(Difference between revisions)
| (12 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
==E. coli== | ==E. coli== | ||
| - | + | ===Construction of RSR+Leader Array=== | |
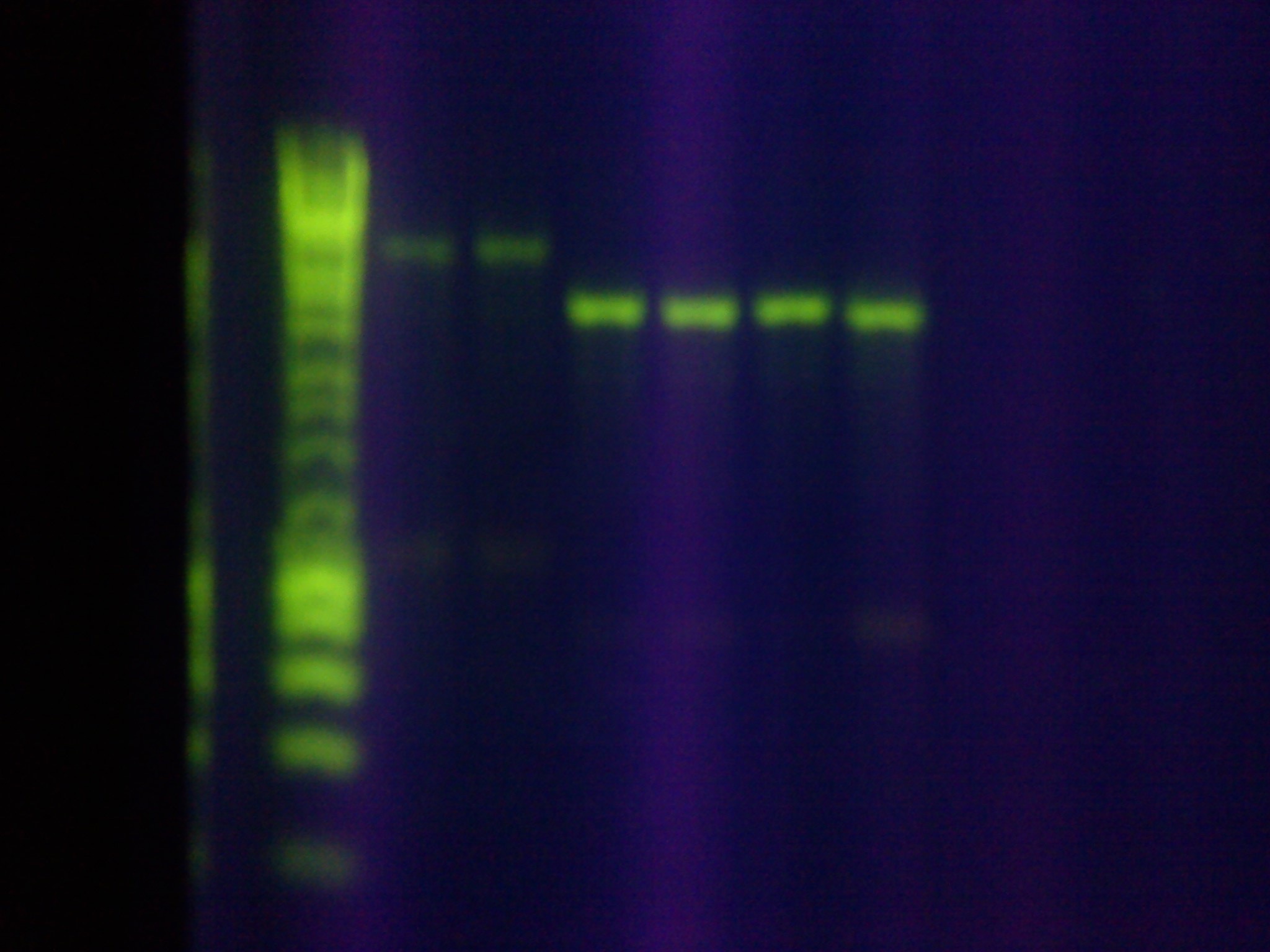
| - | :*Gel results: | + | :*Gel results: Correct size for the RSR+Leader Array in the 6th well. Confirmed with [http://sequencing.biodesign.asu.edu/ DNASU] sequencing. |
| - | [[Image]] | + | [[Image: ASU_RSRLeader.jpg|400px|center]] |
:*Sequencing verification showed a 99% base pair match for "Seq1" and the MG1655 leader sequence, confirming the successful assembly of the array | :*Sequencing verification showed a 99% base pair match for "Seq1" and the MG1655 leader sequence, confirming the successful assembly of the array | ||
| - | |||
| - | + | ||
| + | ===Isolation of Cas genes=== | ||
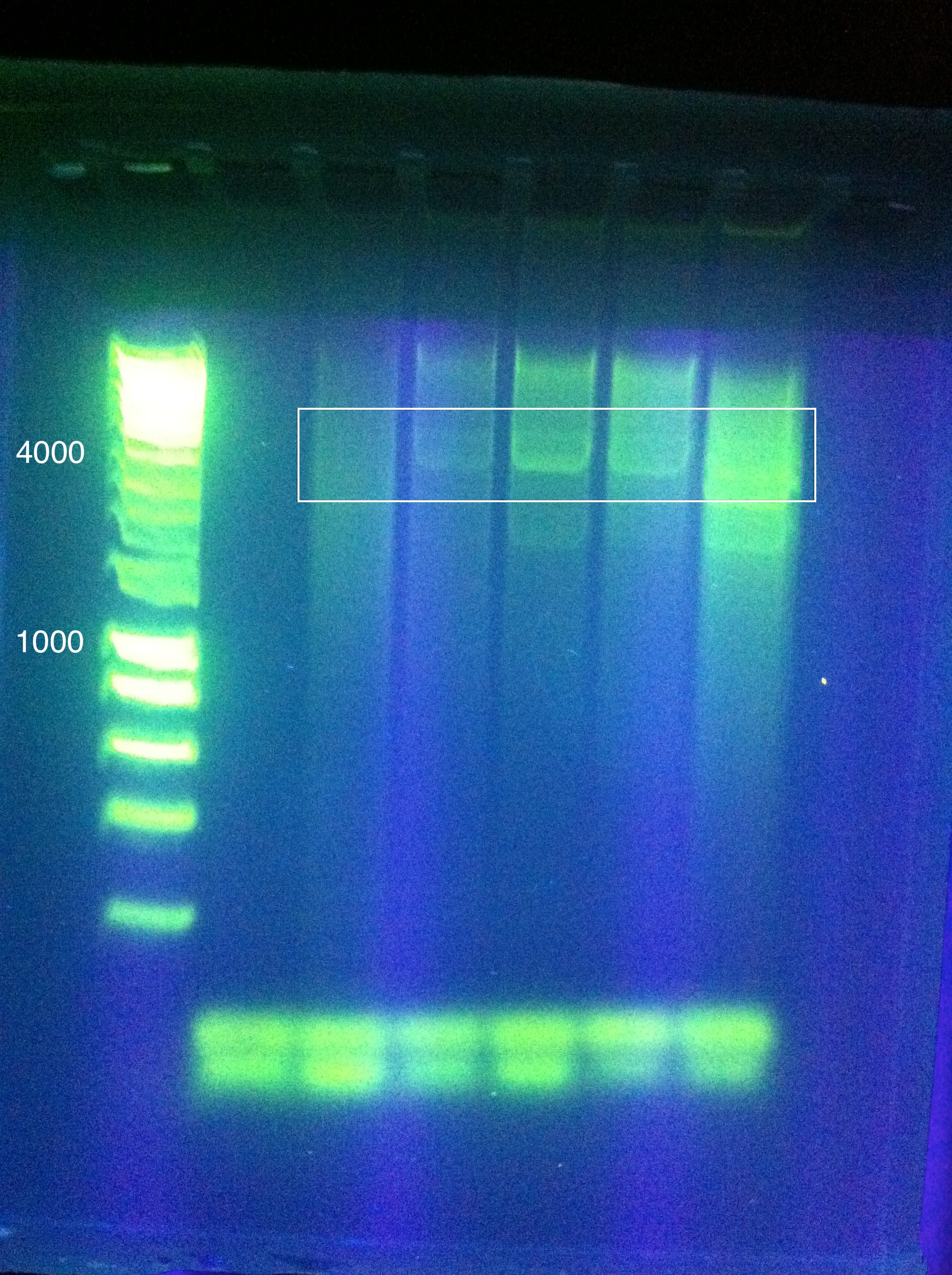
:*CasABCDE was unsuccessful, though we came close: | :*CasABCDE was unsuccessful, though we came close: | ||
| - | [[Image: | + | [[Image: ASU_719_casABCDE.jpg|400px|center]] |
:*Cas3 was moderately successful, but unconfirmed via sequencing: | :*Cas3 was moderately successful, but unconfirmed via sequencing: | ||
| - | [[Image: ASU_725_gel_cas3.jpeg|400px]] | + | [[Image: ASU_725_gel_cas3.jpeg|400px|center]] |
| + | |||
| + | ===Assembly of Leader+RSR into pRSF Duet=== | ||
| + | :*Assembled on 9/28/2011. | ||
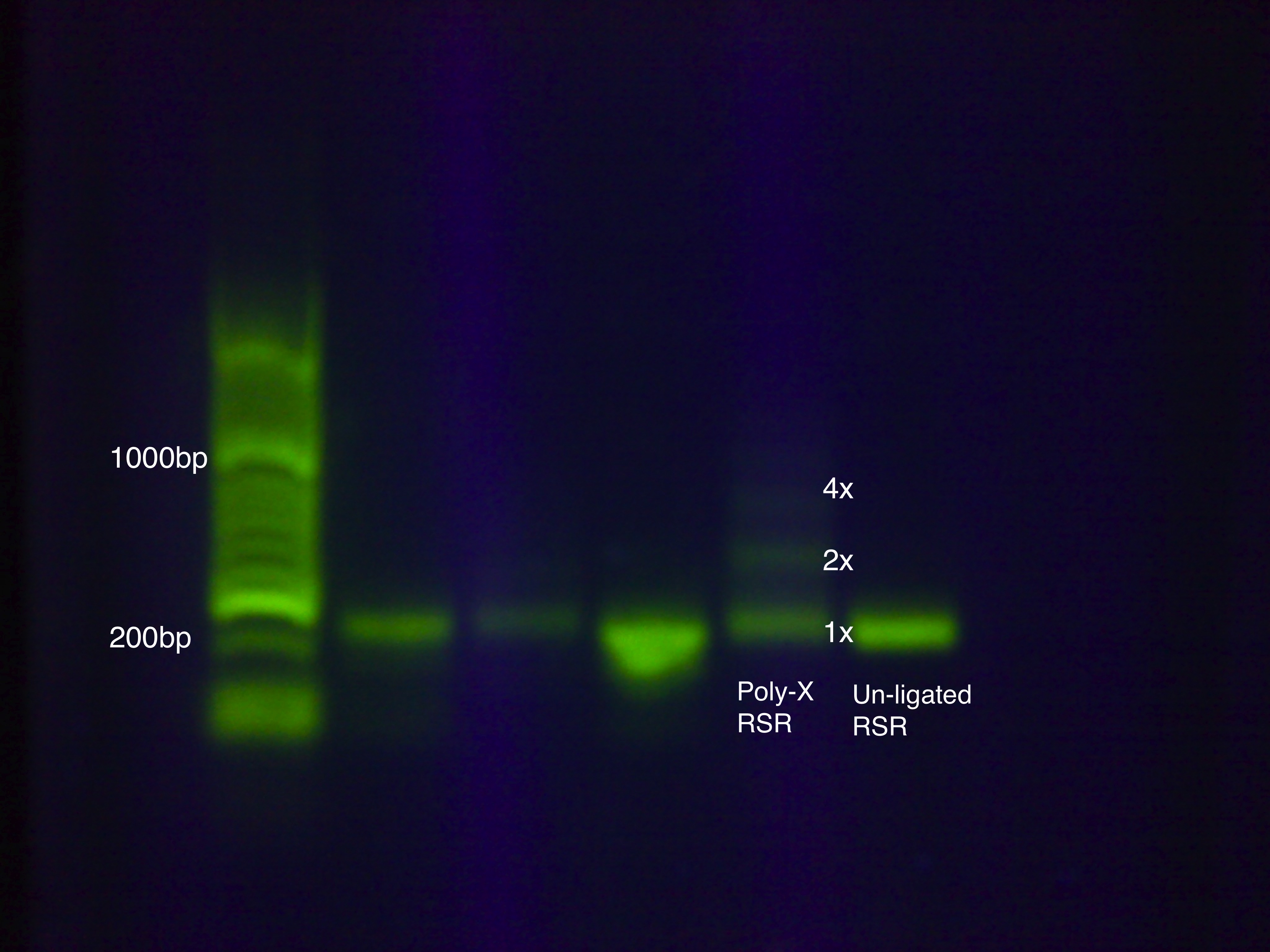
| - | + | ===[https://2011.igem.org/Team:Arizona_State/Lab/Protocols/Assembly#Poly Poly-X Ligation]=== | |
| - | : | + | We devised a new assembly method, the Poly-X Ligation, for RSR construction and tested it on an RSR construct with GFP. |
| + | [[image: Poly-X_RSR.jpg|400px|center]] | ||
| - | + | ===Contingency Test=== | |
:*Standardized competent cells | :*Standardized competent cells | ||
:*Plate results: | :*Plate results: | ||
| Line 27: | Line 31: | ||
==B. halodurans== | ==B. halodurans== | ||
| - | + | ===Construction of RSR Array=== | |
:*We constructed a 1x Repeat-Spacer-Repeat array by ligating our "RA" (Spacer-Repeat) sequence to our "RB" (Repeat) sequence | :*We constructed a 1x Repeat-Spacer-Repeat array by ligating our "RA" (Spacer-Repeat) sequence to our "RB" (Repeat) sequence | ||
| - | |||
| - | + | ===Isolation of cmr genes=== | |
:*Our construct requires the isolation of 6 genes, Cmr1-6, that are all located on a single locus. | :*Our construct requires the isolation of 6 genes, Cmr1-6, that are all located on a single locus. | ||
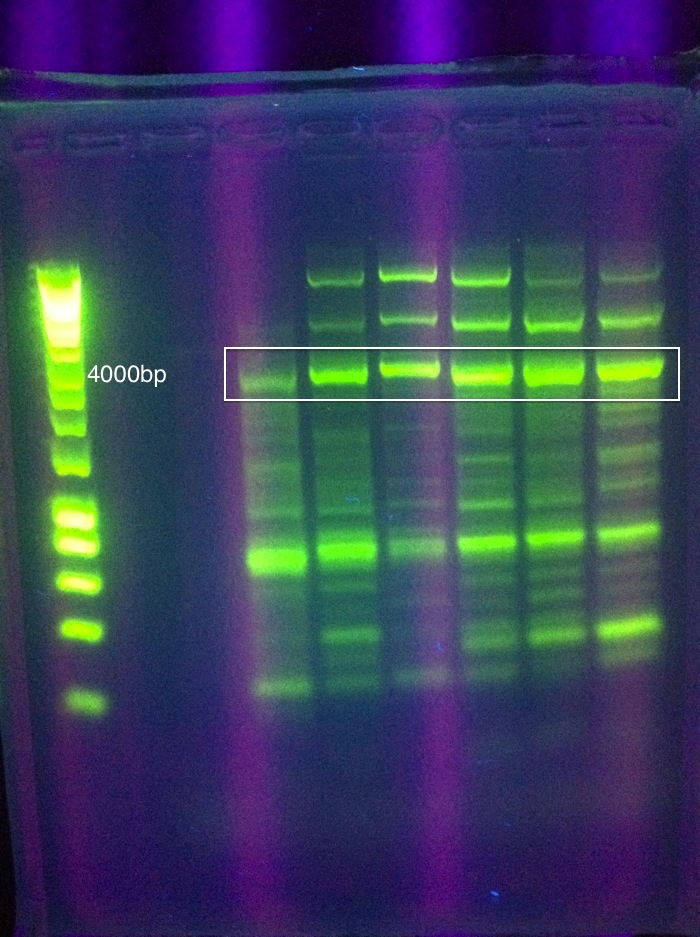
| - | :*Gel results | + | :*Gel results (CMR success): |
| - | [[Image: ASU_716_cmr_success.jpg|400px| | + | [[Image: ASU_716_cmr_success.jpg|400px|center]] |
| - | + | ===Assembly into pRSF Duet=== | |
==L. innocua== | ==L. innocua== | ||
| - | + | ===Construction of RSR Array=== | |
| - | :*This construction of the RSR array involved the ligation of two customized BioBricks ( | + | :*This construction of the RSR array involved the ligation of two customized BioBricks ([[Team:Arizona State/Notebook/Sequences|sequences]]), which can be done simultaneously using this [[Team:Arizona State/Lab/Protocols/Assembly|protocol]]. |
| - | + | ||
| - | [[ | + | |
| - | + | ===Cas gene=== | |
| - | :*The L. innocua CRISPR system utilizes three CRISPR-associated genes | + | :*The L. innocua CRISPR system utilizes three CRISPR-associated genes: Cas1, Cas2, and Cas9. |
:*For our streamlined construct, we needed to isolate only Cas9, which is approximately 4kb in length. | :*For our streamlined construct, we needed to isolate only Cas9, which is approximately 4kb in length. | ||
:*We PCR amplified Cas9 with the adjacent trans-encoded CRIPSR RNA region (tracrRNA), which is necessary for the formation of mature CRISPR RNA. | :*We PCR amplified Cas9 with the adjacent trans-encoded CRIPSR RNA region (tracrRNA), which is necessary for the formation of mature CRISPR RNA. | ||
| - | |||
| - | |||
| - | + | ===Assembly into pRSF Duet=== | |
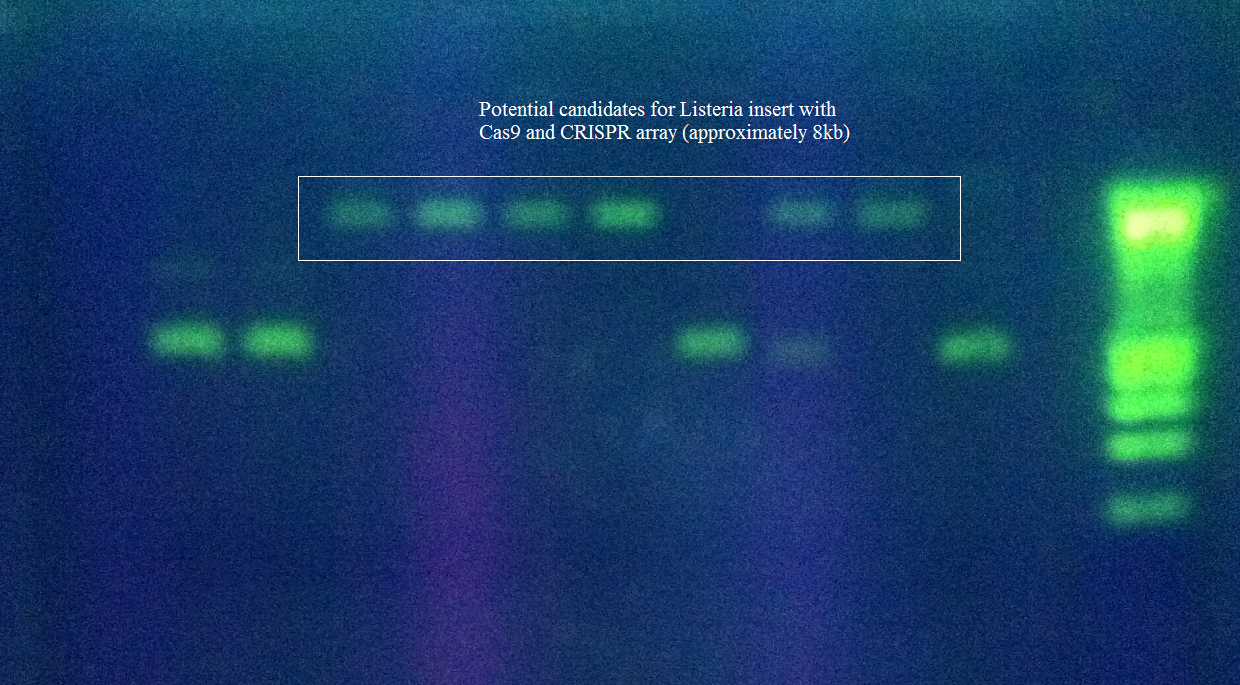
:*Cas9+tracrRNA array has been ligated into pRSF Duet | :*Cas9+tracrRNA array has been ligated into pRSF Duet | ||
| - | [[Image]] | + | [[Image: ASU_Listeria_Gel_9-28.PNG|600px|center]] |
==Other== | ==Other== | ||
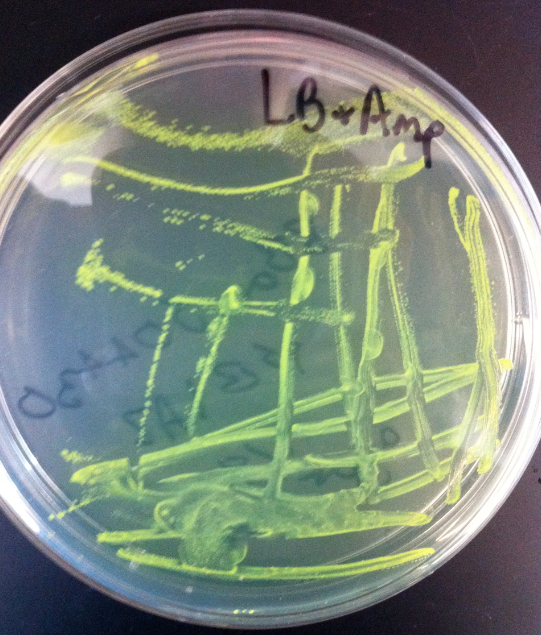
*Construction of constitutive GFP plasmid by ligation of Bba_E0840 with Bba_(promoter) | *Construction of constitutive GFP plasmid by ligation of Bba_E0840 with Bba_(promoter) | ||
:*Success was marked by green colonies | :*Success was marked by green colonies | ||
| - | [[Image]] | + | [[Image: ASU_gfpconstruct.jpg|400px|center]] |
}} | }} | ||
Latest revision as of 04:24, 29 September 2011
|
|
E. coliConstruction of RSR+Leader Array
Isolation of Cas genes
Assembly of Leader+RSR into pRSF Duet
Poly-X LigationWe devised a new assembly method, the Poly-X Ligation, for RSR construction and tested it on an RSR construct with GFP. Contingency Test
*Recorded OD600 = raw absorbance * 3.2
**Weighted Colony Count = (# of Colonies)/(mean OD600 of competency prep) B. haloduransConstruction of RSR Array
Isolation of cmr genes
Assembly into pRSF DuetL. innocuaConstruction of RSR ArrayCas gene
Assembly into pRSF Duet
Other
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"