Team:Arizona State/Notebook/June
From 2011.igem.org
(Difference between revisions)
| (7 intermediate revisions not shown) | |||
| Line 8: | Line 8: | ||
== Thursday, June 2 == | == Thursday, June 2 == | ||
* Meeting with Dr. Chang and her grad students from the School of Life Sciences to explain iGEM project. | * Meeting with Dr. Chang and her grad students from the School of Life Sciences to explain iGEM project. | ||
| - | :* | + | :* Single sided synthesis with phosphotase direction? |
* Made amp plates: | * Made amp plates: | ||
:* 100 mg / ml amp | :* 100 mg / ml amp | ||
| Line 18: | Line 18: | ||
* Incubated and shook liquid culture overnight | * Incubated and shook liquid culture overnight | ||
== Friday, June 3 == | == Friday, June 3 == | ||
| - | * | + | * Confirmed placement of synthesis order |
| - | * | + | * Placed second large order for necessary laboratory materials |
| - | * | + | * Met with Barrett funding advisor |
| - | * | + | * Met with Jon, grad student from Misra's lab |
| - | * | + | * Lab: |
| - | :* | + | :* Resuspended part E0840 from well following parts registry protocol |
| - | :* | + | :* Followed "competent cells and chemical transformation procedure for DH5 alpha": |
| - | ::* | + | ::* Made 20mM concentration MgCl2 in shaken cells from yesterday |
| - | ::* | + | ::* Shook for 2 hours in 37 C room |
== Saturday, June 4 == | == Saturday, June 4 == | ||
| - | * | + | * Made 500 mL SOC following OpenWetWare protocol |
| - | * | + | * Transformed resuspended DNA into E. Coli |
| - | :* | + | :* Followed transformation protocol, but did not use water bath |
== Monday, June 6 == | == Monday, June 6 == | ||
| - | * | + | * Transformed cells from Saturday, June 4th did not grow yet |
| - | * | + | * Test if competency procedure killed cells using following procedure: |
| - | :* | + | :* Make LB= |
| - | * | + | * Repeat transformation using water bath instead of heat block: |
| - | :* | + | :* Thaw competent cells on ice |
:* 50 ul cells + 1 ul resuspended DNA, on ice for 30 minutes | :* 50 ul cells + 1 ul resuspended DNA, on ice for 30 minutes | ||
| - | :* | + | :* Heat shock cells at 42 in a water bath for 60 seconds |
| - | :* | + | :* Incubate on ice, 5 min |
| - | :* | + | :* Add 100 ul SOC to cells |
| - | :* | + | :* Shake at 37 C for 2 hours (11 am - 1 pm) |
| - | :* | + | :* Plate 20 ul, 200 ul (2 plates) |
| - | :* | + | :* Incubate overnight |
== Tuesday, June 7 == | == Tuesday, June 7 == | ||
| - | * | + | * Amp plates did not grow |
| - | * | + | * Competent cells plated without amp grew |
| - | * | + | * New transformation using 2 different parts conducted did not work with either part: |
:* BBa_E0840 | :* BBa_E0840 | ||
:* BBa_E0240 | :* BBa_E0240 | ||
| - | * | + | * Control onto non amp plate to test if transformation killed cells showed that the cells were still viable |
== Wednesday, June 8 == | == Wednesday, June 8 == | ||
| - | * | + | * Autoclaved lab materials to be sterilized |
| - | * | + | * Made 200 ml new SOB, 50 ml SOC |
| - | * | + | * New transformation protocols: |
| - | :* | + | :* Top10 chemically competent E. Coli from biodesign |
| - | :* | + | :* Part: BBa_E0840 |
| - | :* | + | :* Using top10 protocol |
| - | :* | + | :* Plates: # 4 50ul, 5 150ul |
* MG1655 plated from plate # 2: | * MG1655 plated from plate # 2: | ||
| - | :* | + | :* Plate: # 1 |
| - | :* | + | :* From: 6-2 plate MG1655 |
| - | * | + | * Overnight culture: |
| - | :* | + | :* From plate # 3 |
== Thursday, June 9 == | == Thursday, June 9 == | ||
* Xiao introduced 3 grad students who can offer advice/assistance throughout the project | * Xiao introduced 3 grad students who can offer advice/assistance throughout the project | ||
:* We will meet with them (likely Thursdays @ 10am) to update them on our progress | :* We will meet with them (likely Thursdays @ 10am) to update them on our progress | ||
| - | * | + | * Yesterday's plates: |
| - | :* #4, 5 have colonies but no glow with UV- no promoter in biobrick part | + | :* #4, 5 have colonies but no glow with UV - no promoter in biobrick part |
| - | ::* | + | ::* Need to add in a promoter |
* Today: | * Today: | ||
| - | * | + | * Add in promoter for GFP construct |
| - | :* | + | :* Constitutive promoter: |
| - | ::* | + | ::* Part: BBa_J23101 |
| - | :* | + | :* Use Knight restriction protocol |
| - | ::* | + | ::* Cut promoter BBa_J23101 with ECORI, SPEI |
| - | ::* | + | ::* Cut GFP generator BBa_e0840 with ECORI, XHOI |
::* DNA extraction- use "ethanol precipitation of nucleic acid" procedure | ::* DNA extraction- use "ethanol precipitation of nucleic acid" procedure | ||
| - | :* Ligate | + | :* Ligate restriction products |
| - | :* Transform | + | :* Transform ligation products |
:* Create stock of competent cells | :* Create stock of competent cells | ||
* Order Top10 cells (what strain are these?) | * Order Top10 cells (what strain are these?) | ||
| - | * Make glycerol stock of | + | * Make glycerol stock of BioBrick |
* Jon/Misra procedure for competent cells and transformation: | * Jon/Misra procedure for competent cells and transformation: | ||
| - | :* | + | :* Overnight culture from previous: |
| - | ::* | + | ::* Diluted 1 to 50 |
| - | ::* | + | ::* Shook 1 hr in 37 C room |
== Friday, June 10 == | == Friday, June 10 == | ||
| - | * | + | * Lab today: |
:* 2 competency procedures (Jon, CCMB80) | :* 2 competency procedures (Jon, CCMB80) | ||
| - | :* 3 transformation procedures ( | + | :* 3 transformation procedures (Jon, CCMB80, top10 from biodesign) |
:* 12 plates made (see lab notebook) | :* 12 plates made (see lab notebook) | ||
* Autoclaved glass test tubes | * Autoclaved glass test tubes | ||
| - | * | + | * Dan demonstrated to lab members how to make glycerol stocks |
| - | * Bought competent cells | + | * Bought top10 competent cells |
| - | + | ||
* Got account set up (still need to create Sunrise account) | * Got account set up (still need to create Sunrise account) | ||
:* Got Xiao refunded | :* Got Xiao refunded | ||
| - | * Met | + | * Met with James Alling, who is a JD-PhD interested in helping us out |
| - | :* He is very good at speaking and could help | + | :* He is very good at speaking and could help with presentation later |
:* Very attracted to promoting big picture of project | :* Very attracted to promoting big picture of project | ||
* Kylie determined new primers after noticing that we don't need Cas 1,2,3 for our natural cas construct (from "structural basis for CRISPR") | * Kylie determined new primers after noticing that we don't need Cas 1,2,3 for our natural cas construct (from "structural basis for CRISPR") | ||
| - | :* This brings down | + | :* This brings Cas construct size down to a total of 3.8kb instead of over 5kb |
| - | :* We will try both ways, see if cas 1 | + | :* We will try both ways, and see if cas 1 and 2 do anything interesting |
| - | * primers for | + | * We are considering getting primers for each individual Cas gene (Cas A, Cas B, etc...) |
* Biobasic is taking twice as long as they advertised (no DNA until june 20?) | * Biobasic is taking twice as long as they advertised (no DNA until june 20?) | ||
| - | :* From now on we will go through IDT | + | :* From now on we will go through IDT due to slow turnaround from BioBasic |
| - | :* | + | :* We contacted BioBasic about discount, got a synthesis price reduction |
== Saturday, June 11 == | == Saturday, June 11 == | ||
HAPPY 22ND BIRTHDAY KEITH! | HAPPY 22ND BIRTHDAY KEITH! | ||
| + | <br> | ||
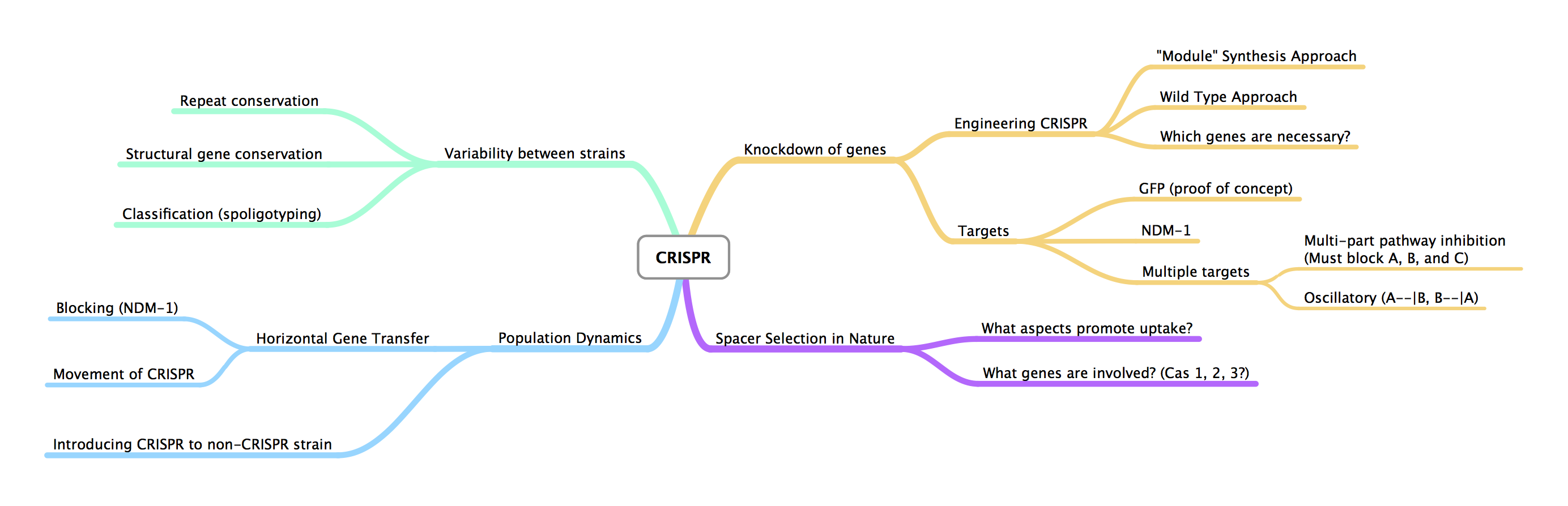
| + | *Created idea web for CRISPR in MindNode | ||
| + | [[Image:ASU_CRISPR_Mindnode.pdf|800px]] | ||
== Tuesday, June 14 - Friday, June 17 == | == Tuesday, June 14 - Friday, June 17 == | ||
| - | * Synthetic Biology 5.0 conference | + | * [https://2011.igem.org/Team:Arizona_State/Outreach/Events Synthetic Biology 5.0 conference] |
== Monday, June 20 == | == Monday, June 20 == | ||
* today: | * today: | ||
| - | :* overnight culture x 2 (LB) for DNA | + | :* Prepared overnight culture x 2 (LB) for DNA miniprepping |
| - | :* overnight culture x 2 (LB, SOC) for culture | + | :* Prepared overnight culture x 2 (LB, SOC) for culture |
::* CCMB80 | ::* CCMB80 | ||
| - | * | + | * Tomorrow: |
| - | :* genome prep | + | :* Carry out genome prep for K12 genome from MG1655 |
| - | :* | + | :* Begin PCR amplification of Cas genes |
| - | :* competent cells | + | :* Create and test competent cells |
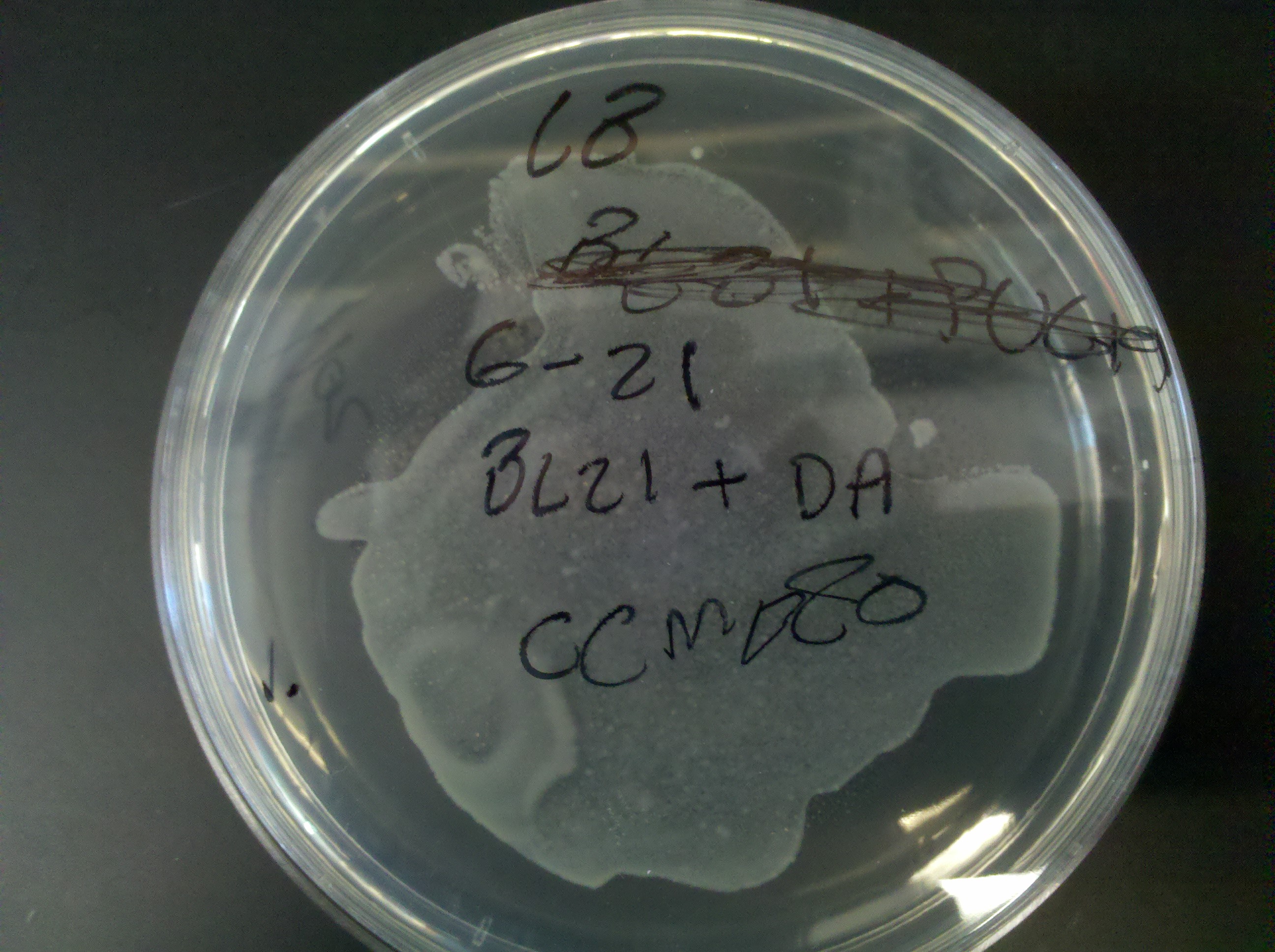
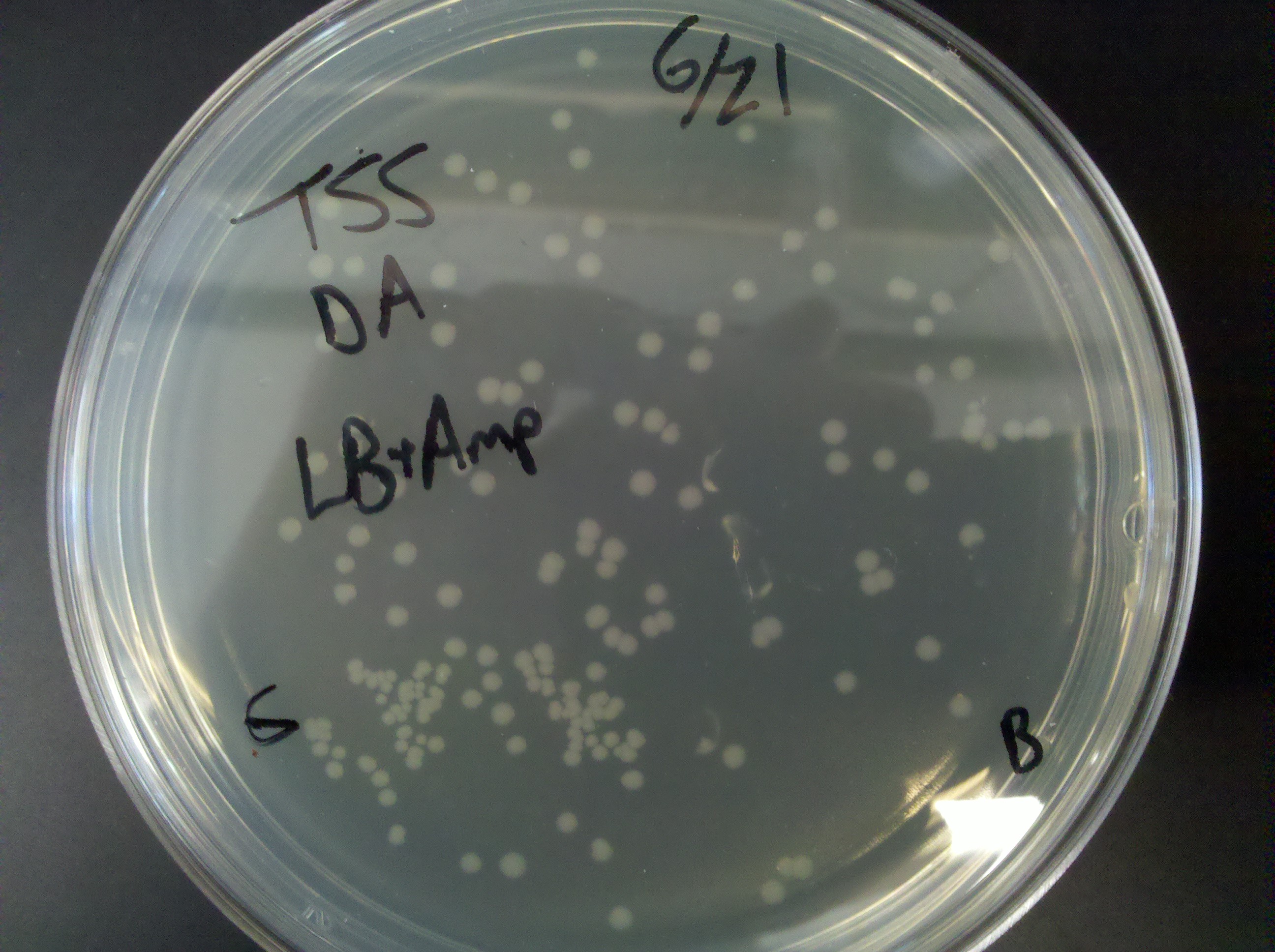
== Tuesday, June 21 == | == Tuesday, June 21 == | ||
| - | * | + | * Competency information: |
:* CCMB80 w/ BL21 cells | :* CCMB80 w/ BL21 cells | ||
| - | ::* | + | ::* Ruben and Ethan carried out this competency procedure |
| - | ::* | + | ::* Made 250 ml SOB |
| - | ::* 9 210ul tubes | + | ::* 9 210ul tubes placed into -80 C fridge |
::* plates made: | ::* plates made: | ||
:::* 1. LB, transformation: DA | :::* 1. LB, transformation: DA | ||
| Line 132: | Line 134: | ||
:::* 3. LB + amp, transformation: PUC19 | :::* 3. LB + amp, transformation: PUC19 | ||
:* TSS procedure w/ BL21 cells | :* TSS procedure w/ BL21 cells | ||
| - | ::* | + | ::* Madeline, Juan, and Keith carried out this competency procedure |
::* plates made: | ::* plates made: | ||
:::* 4. LB + amp, PUC19, burned | :::* 4. LB + amp, PUC19, burned | ||
| Line 139: | Line 141: | ||
:::* 7. LB + amp, DA | :::* 7. LB + amp, DA | ||
:::* 8. LB + +amp, DA | :::* 8. LB + +amp, DA | ||
| - | :* NEB top10: | + | :* NEB top10 competency test: |
| - | ::* | + | ::* Plates made: |
:::* 9. LB + amp, PU19 | :::* 9. LB + amp, PU19 | ||
:::* 10. LB + amp, PUC19 | :::* 10. LB + amp, PUC19 | ||
:::* 11. LB + amp, DA | :::* 11. LB + amp, DA | ||
| - | * | + | * Genome prep: |
| - | :* | + | :* Carried out by Nisarg |
| - | * First PCR overnight of CAS genes | + | * First PCR conducted overnight of attempting to amplify CAS genes A-E and 3 |
== Wednesday, June 22 == | == Wednesday, June 22 == | ||
| - | * | + | * Made new LB + amp stock |
| - | * | + | * Made 200 ml LB + amp broth |
| - | * plates | + | * Results from plates made yesterday: |
| - | :* | + | :* Ampicillin stock integrity in question |
: 1: normal growth (no distinct colonies) <br> [[Image: ASU_622_1.jpg|400px]] | : 1: normal growth (no distinct colonies) <br> [[Image: ASU_622_1.jpg|400px]] | ||
: 2: colonies | : 2: colonies | ||
| Line 163: | Line 165: | ||
: 10: very heavy colonies <br> [[Image: ASU_622_10.jpg|400px]] | : 10: very heavy colonies <br> [[Image: ASU_622_10.jpg|400px]] | ||
: 11: light colonies | : 11: light colonies | ||
| - | * | + | * Overnight cultures made of 2, 6, 7, 8, 11 (3 each in LB + amp broth) |
| - | * gel of | + | * Ran a gel of PCR product |
| - | * | + | * New plates: |
: 1. CCMB80, LB, PUC19 | : 1. CCMB80, LB, PUC19 | ||
: 2. CCMB80, LB, DB | : 2. CCMB80, LB, DB | ||
| Line 190: | Line 192: | ||
: 23. TSS, LB + amp, DB | : 23. TSS, LB + amp, DB | ||
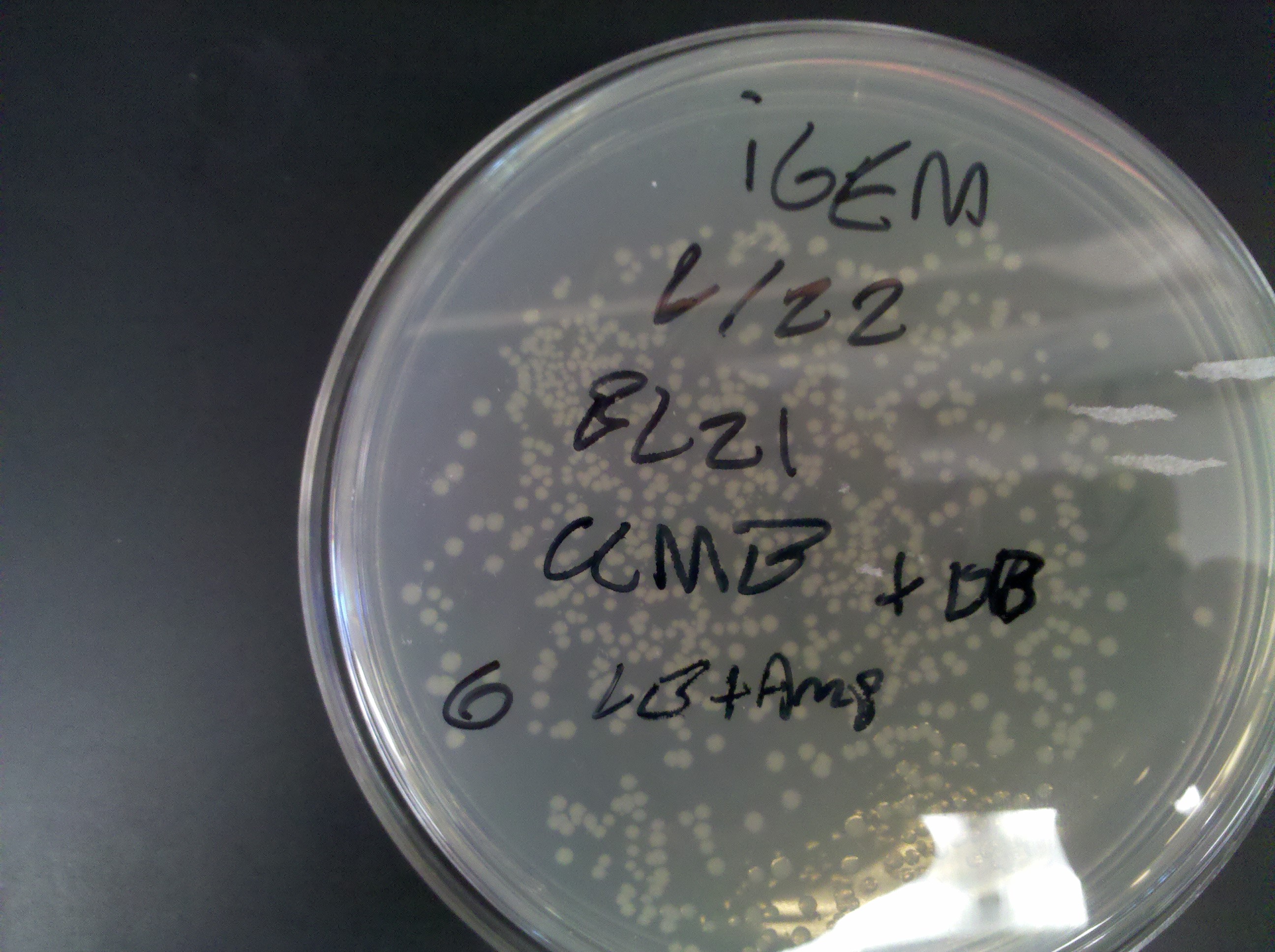
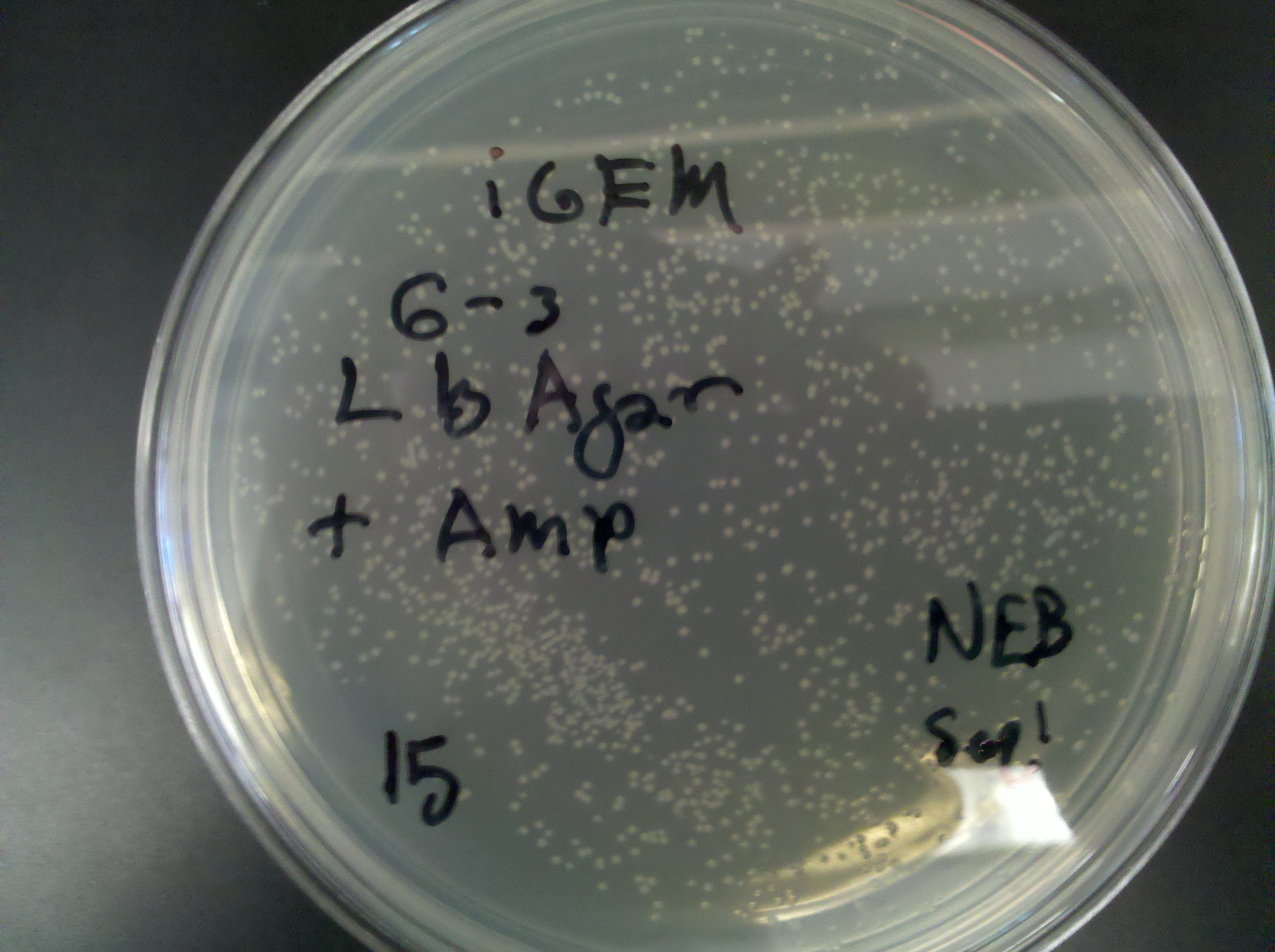
== Thursday, June 23 == | == Thursday, June 23 == | ||
| - | * | + | * Plates from yesterday worked completely as expected |
Plate 6: <br> [[Image: ASU_623_6.jpg|400px]] <br> | Plate 6: <br> [[Image: ASU_623_6.jpg|400px]] <br> | ||
Plate 15: <br> [[Image: ASU_623_15.jpg|400px]] | Plate 15: <br> [[Image: ASU_623_15.jpg|400px]] | ||
| - | * | + | * Overnight culture in amp grew |
| - | * | + | * Today: |
| - | :* | + | :* Glycerol stock made of DA |
| - | :* | + | :* Overnight cultures made of DB, sEQ1 from plates |
| - | :* gel of PCR from last night | + | :* Ran gel of PCR from last night |
:* DNA extraction 2x (elution)- verified using nanodrop, did not get enough to be successful | :* DNA extraction 2x (elution)- verified using nanodrop, did not get enough to be successful | ||
| - | :* | + | :* Another overnight PCR using different settings |
| - | :* | + | :* Designed new primers for casA-E + cas3 |
== Friday, June 24 == | == Friday, June 24 == | ||
| - | * gel from | + | * Ran gel from PCR carried out last night |
| - | :* | + | :* Still doesn't work! |
| - | * | + | * DNA extraction using spin method from Miniprep protocol(DB, SEQ1) |
| - | :* | + | :* Still doesn't work! |
| - | ::* went through hassle of ordering new | + | ::* We went through hassle of ordering new Cas primers from IDT |
| - | : | + | : Ordered a pair of primers for each Cas gene - this way we can customize and perhaps PCR out in sections |
== Saturday, June 25 == | == Saturday, June 25 == | ||
| - | * | + | * Transformations of DA, DB, and Seq1 into the BioBrick ampR vector (pSB1A3) into TSS and NEB cells was successful |
| - | * | + | * Made overnight liquid culture to miniprep tomorrow |
== Sunday, June 26 == | == Sunday, June 26 == | ||
| - | * | + | * Made LB amp plates |
| - | * restriction digest | + | * Conducted restriction digest... |
| - | :* wrong enzymes! used EX and EP instead of EX and ES | + | :* We used the wrong enzymes! used EX and EP instead of EX and ES |
| - | * | + | * Ran gel on previous PCR |
| - | :* | + | :* Didn't linearize plasmid before running results on a gel |
== Monday, June 27 == | == Monday, June 27 == | ||
| - | * | + | * We identified the Top 10 lab techniques to learn and love |
| - | :* | + | :* Dan emphasized that we need to be independent and know these! |
| - | * | + | * Redid restriction digest |
| - | :* | + | :* Two methods for restriction: Ginkgo bioworks (two bricks into desired plasmid) and traditional (EX and ES) |
:* DA: ES, EX | :* DA: ES, EX | ||
:* Seq1: ES, XP | :* Seq1: ES, XP | ||
:* PSB1A3: EX, EP | :* PSB1A3: EX, EP | ||
| - | * | + | * Some Gel errors |
: 1) did not let gel dry completely before removing comb | : 1) did not let gel dry completely before removing comb | ||
: 2) too much voltage caused gel deformation | : 2) too much voltage caused gel deformation | ||
| - | * | + | * Ran out of PSB1A3 |
| - | :* | + | :* Lesson learned: don't use it directly! must grow it up first |
| - | :* | + | :* Ordered more from iGEM HQ |
| - | * | + | * Cultured B. Halodurans |
| - | :* rehydrated and let culture overnight in tryptic soy broth | + | :* We rehydrated cells and let culture grow overnight in tryptic soy broth |
| - | * overnight | + | * Made overnight cultures of Seq1, DA, DB, and E0840 |
* Overall message: Not a great day in terms of results, but many tough lessons learned. | * Overall message: Not a great day in terms of results, but many tough lessons learned. | ||
== Tuesday, June 28 == | == Tuesday, June 28 == | ||
| - | * | + | * B. halodurans developments |
| - | :* from overnight culture | + | :* Cells retrieved from overnight culture |
| - | ::* | + | ::* Made 4 plates on tryptic soy media, as well as 7 more tryptic soy plates |
| - | ::* glycerol stock | + | ::* Made one glycerol stock |
| - | :* Genomic prep + PCR using primers R1 and R2 | + | :* Conducted Genomic prep + PCR using primers R1 and R2 |
* Nanodrop new record! 220ng/ul template DNA | * Nanodrop new record! 220ng/ul template DNA | ||
* "Ode to Trinette" Haiku by Joseph Flay | * "Ode to Trinette" Haiku by Joseph Flay | ||
| Line 246: | Line 248: | ||
: Trinette, you are so thermal | : Trinette, you are so thermal | ||
: Thanks for the fun times | : Thanks for the fun times | ||
| - | * | + | * Conducted miniprep of Seq1, DA, DB, E0840 |
| - | * Nanodrop results (see Kylie's notebook) | + | :* Nanodrop results (see Kylie's notebook) |
| - | * | + | * Further restriction digests |
| - | :* Made " | + | :* Made "restriction supermix" of water, BSA, NEB4 (1x, enough for 30 digests) |
:* Seq 1: ES, EX | :* Seq 1: ES, EX | ||
:* DA: ES, EX | :* DA: ES, EX | ||
:* DB: ES, EX | :* DB: ES, EX | ||
:* E0840: EP | :* E0840: EP | ||
| - | :* | + | :* Made and ran a large gel |
:* Problem: used wrong hyperladder (used I instead of II) | :* Problem: used wrong hyperladder (used I instead of II) | ||
* Successfully isolated: Seq 1 (ES), Seq 1 (EX), DB (EX), E0840 (insert), E0840 (vector) | * Successfully isolated: Seq 1 (ES), Seq 1 (EX), DB (EX), E0840 (insert), E0840 (vector) | ||
| Line 261: | Line 263: | ||
* Replated BL21DE3 x 1 and MG1655 x 1 on LB Agar | * Replated BL21DE3 x 1 and MG1655 x 1 on LB Agar | ||
* Moved plates from 4 degree room to small fridge in lab because they are fixing the room tomorrow (and got rid of some old plates) | * Moved plates from 4 degree room to small fridge in lab because they are fixing the room tomorrow (and got rid of some old plates) | ||
| - | * Took inventory (mostly) | + | * Took lab inventory (mostly) |
| - | * | + | * Made more overnight cultures: |
:* B. halodurans x 1 | :* B. halodurans x 1 | ||
* Other notes: Paul Johnson sent us a nice message basically saying that as long as we can justify it, iGEM is here to stay | * Other notes: Paul Johnson sent us a nice message basically saying that as long as we can justify it, iGEM is here to stay | ||
| Line 268: | Line 270: | ||
*: We also talked about getting FURI and SOLUR funding for next year's team. | *: We also talked about getting FURI and SOLUR funding for next year's team. | ||
*: An REU proposal was discussed, but ultimately abandoned because a majority of the team's students would need to be from outside ASU, which we don't want. | *: An REU proposal was discussed, but ultimately abandoned because a majority of the team's students would need to be from outside ASU, which we don't want. | ||
| - | * Overall: people kept very busy, we worked well in teams, however we need to make sure we are really paying attention to what we do- mistakes cost time! | + | * Overall: people kept very busy, we worked well in teams, however we need to make sure we are really paying attention to what we do - mistakes cost time and money! |
| - | * Tomorrow: plan on ligation, check PCR, run gel for PCR, order primers for halodurans, try restriction of DA again, miniprep and try restriction of RA/RB | + | * Tomorrow: plan on ligation, check PCR results, run a gel for PCR results, order primers for B. halodurans, try restriction of DA again, miniprep and try restriction of RA/RB |
== Wednesday, June 29 == | == Wednesday, June 29 == | ||
| - | * | + | * Plates from last night (see pictures): |
:* LB + AMP + RA | :* LB + AMP + RA | ||
:* LB + AMP + RA | :* LB + AMP + RA | ||
:* LB + AMP + RB | :* LB + AMP + RB | ||
:* LB + AMP + RB | :* LB + AMP + RB | ||
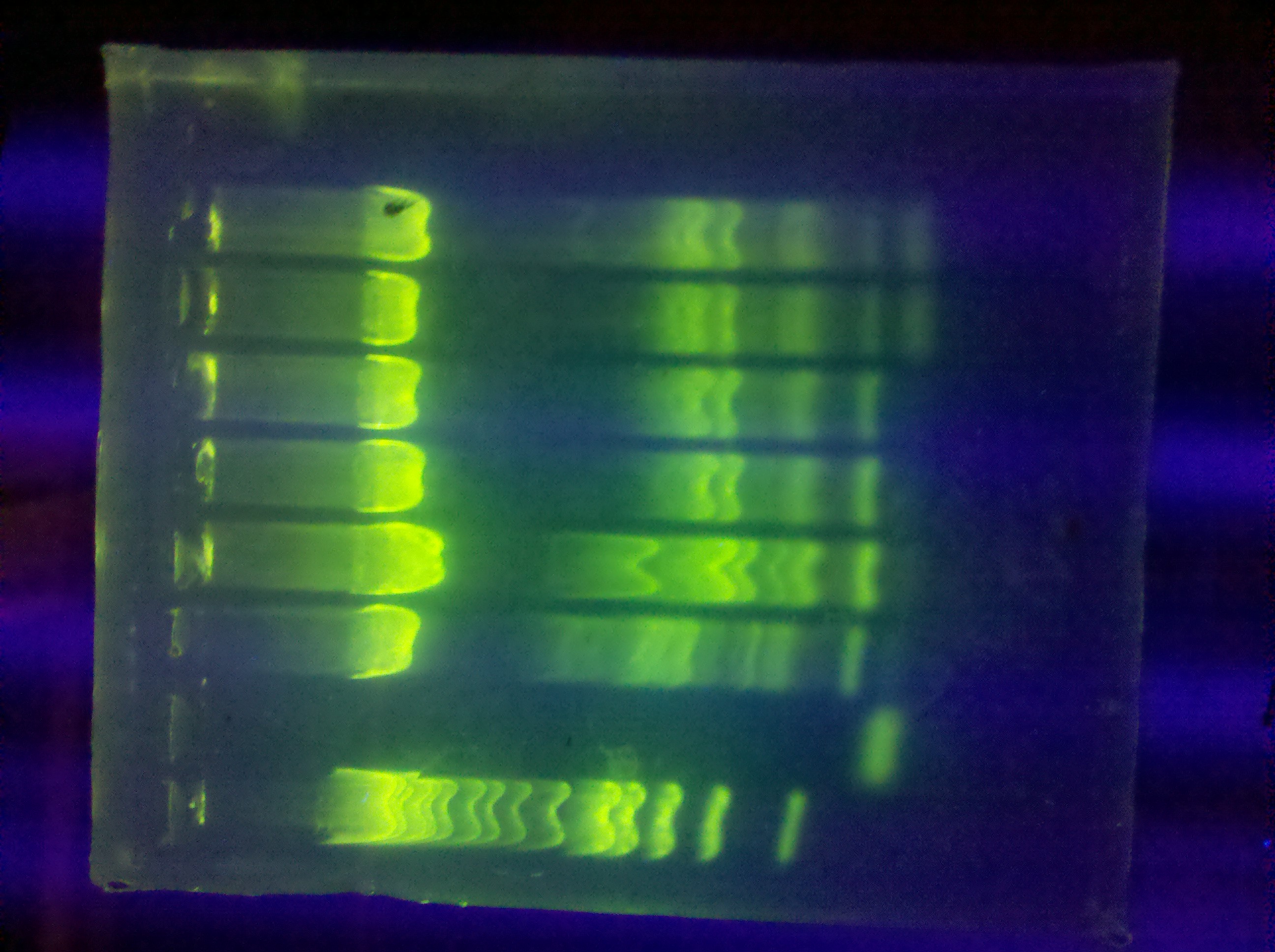
| - | * | + | * Restriction digest of DA, DB (2x) |
| - | * | + | * Run a gel: CMR product from BH PCR |
[[Image: ASU_PCR_gel_629.jpg|400px]] | [[Image: ASU_PCR_gel_629.jpg|400px]] | ||
| - | :* gel extraction, submitted for sequencing | + | :* Conducted gel extraction, submitted extracted DNA for sequencing |
| - | ::* | + | ::* Very low yield (~20 ng/uL) |
== Thursday, June 30 == | == Thursday, June 30 == | ||
* New primers arrived from IDT for second round of attempts at getting the cas genes out of MG1655 | * New primers arrived from IDT for second round of attempts at getting the cas genes out of MG1655 | ||
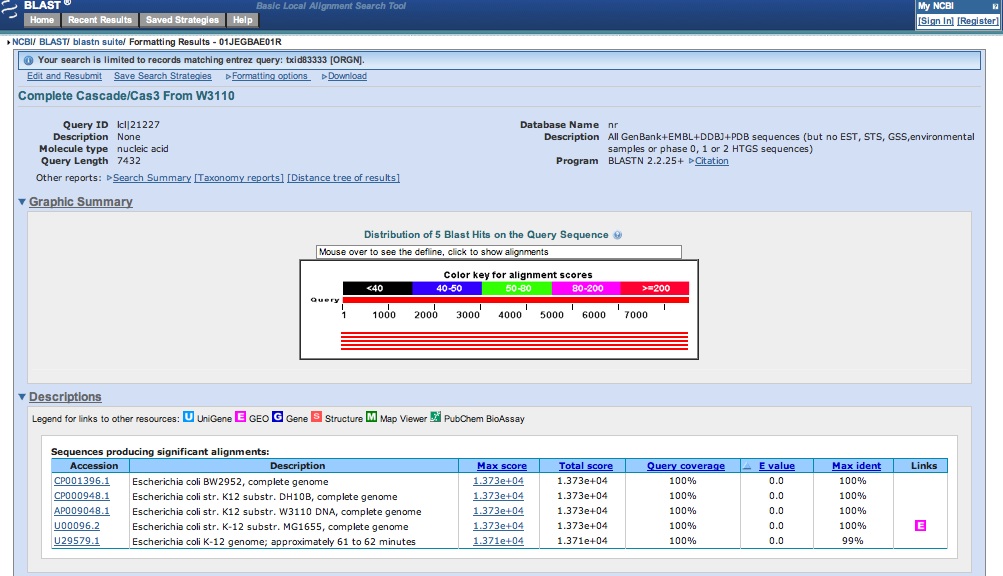
| - | * After successful isolation of what looks like the | + | * After successful isolation of what looks like the CMR genes from Bacillus halodurans, a second attempt was run overnight |
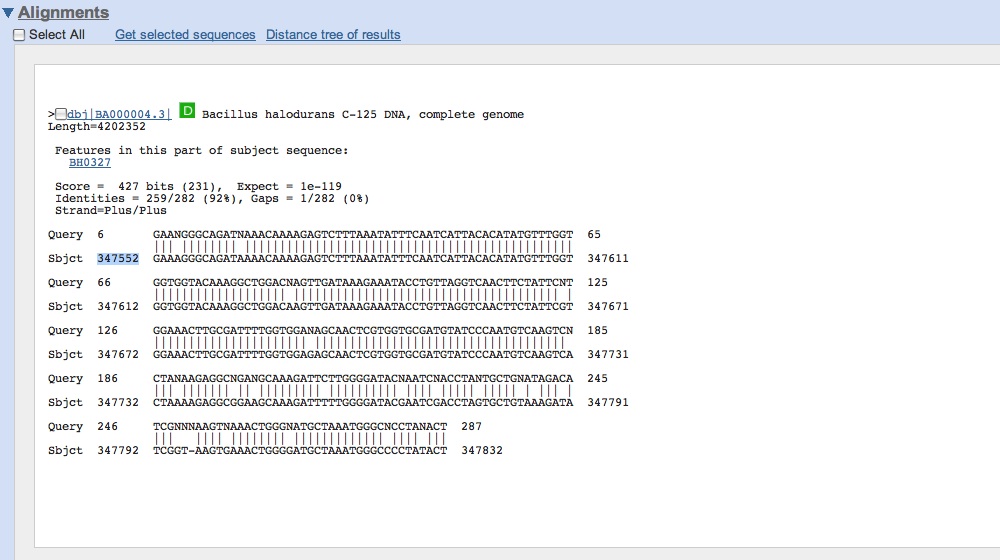
| - | * Got our sequence data from last night for CMR genes: looks like we | + | * Got our sequence data from last night for CMR genes: looks like we successfully amplified CMR! |
<br> [[Image:ASU_630_Cas3.jpg|800px]] | <br> [[Image:ASU_630_Cas3.jpg|800px]] | ||
<br> [[Image:ASU_630_B.H._C125_Genome.jpg|800px]] | <br> [[Image:ASU_630_B.H._C125_Genome.jpg|800px]] | ||
* Gel results: | * Gel results: | ||
:* Cultures of RA/RB grew well | :* Cultures of RA/RB grew well | ||
| - | :* | + | :* Conducted miniprep of RA x2, RB x2 |
| - | * | + | * More restrictions: |
:* DA ES EX (2x) | :* DA ES EX (2x) | ||
:* DB ES XP (2x) | :* DB ES XP (2x) | ||
Latest revision as of 02:05, 29 September 2011
 "
"