Team:Paris Bettencourt/Experiments/pHyperSpank
From 2011.igem.org
Danyel.lee90 (Talk | contribs) |
|||
| (15 intermediate revisions not shown) | |||
| Line 14: | Line 14: | ||
<html> | <html> | ||
<h2>Results</h2> | <h2>Results</h2> | ||
| + | |||
| + | <p>The new biobrick pHS (K606040) was cloned in front of an RFP terminator. The fluorescence was seen under the microscope with strains induced and not induced but IPTG</p> | ||
</html> | </html> | ||
{| border="1" class="wikitable" style="text-align: center;" | {| border="1" class="wikitable" style="text-align: center;" | ||
| - | |E. coli containing Hyperspank +RBS+RFP | + | |E. coli containing Hyperspank +RBS+RFP whithout IPTG at 37°C |
|- | |- | ||
|[[File:minustrans1.jpg|450px|thumb|center|E.coli at 37°C (trans image)]] | |[[File:minustrans1.jpg|450px|thumb|center|E.coli at 37°C (trans image)]] | ||
| - | |[[File:minusfluo1.jpg|450px|thumb|center|E.coli at 37°C | + | |[[File:minusfluo1.jpg|450px|thumb|center|E.coli at 37°C negative control (rfp image)]] |
|- | |- | ||
| - | |[[File:minustrans2.jpg|450px|thumb|center|E.coli | + | |[[File:minustrans2.jpg|450px|thumb|center|E.coli at 37°C (trans image)]] |
|[[File:minusfluo2.jpg|450px|thumb|center|E.coli negative control at 37°C (rfp image)]] | |[[File:minusfluo2.jpg|450px|thumb|center|E.coli negative control at 37°C (rfp image)]] | ||
|} | |} | ||
{| border="1" class="wikitable" style="text-align: center;" | {| border="1" class="wikitable" style="text-align: center;" | ||
| - | |E. coli containing Hyperspank +RBS+RFP with | + | |E. coli containing Hyperspank +RBS+RFP with IPTG at 37°C |
|- | |- | ||
|[[File:plustran1.jpg|450px|thumb|center|E.coli at 37°C (trans image)]] | |[[File:plustran1.jpg|450px|thumb|center|E.coli at 37°C (trans image)]] | ||
|[[File:plusfulo1.jpg|450px|thumb|center|E.coli at 37°C (rfp image)]] | |[[File:plusfulo1.jpg|450px|thumb|center|E.coli at 37°C (rfp image)]] | ||
|- | |- | ||
| - | |[[File:plustrans2.jpg|450px|thumb|center|E.coli | + | |[[File:plustrans2.jpg|450px|thumb|center|E.coli at 37°C (trans image)]] |
| - | |[[File:plusfluo2.jpg|450px|thumb|center|E.coli | + | |[[File:plusfluo2.jpg|450px|thumb|center|E.coli at 37°C (rfp image)]] |
|} | |} | ||
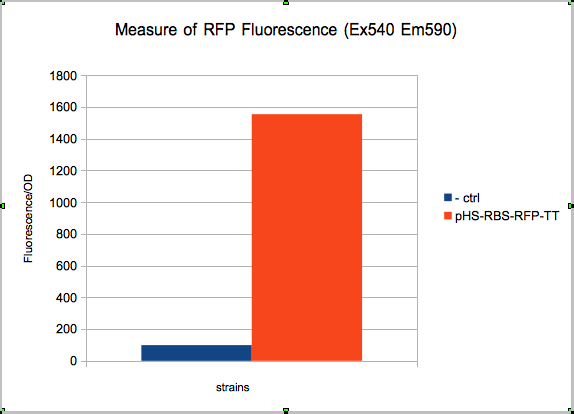
| + | The absolute value of the net fluorescence over OD for induced and non indeuced cells was measured in the TECAN i-control, and plotted below. | ||
| + | [[File:spectrophotometry.jpg|center]] | ||
| + | <html> | ||
| + | <h3>LacI reduces basal expression of IPTG inducible promoter Phyperspank in E.coli (LacIq+).</h3> | ||
| + | <p> Constructs from the <b><a href="https://2011.igem.org/Team:Paris_Bettencourt/Experiments/tRNA_diffusion">tRNA amber system</a></b> were used for these experiments.</p> | ||
| + | <p>The control strain LacI- holds the construct: pT7_RBS_GFP_tt:Pveg_RBS_T7amber_tt:Phyperspank_tRNA_tt</p> | ||
| + | <p>The control strain pT7_GFP:Pveg_T7amber holds the same construct without the tRNA amber. The GFP expression in this strain solely depends on the basal expression level of the T7amber. </p> | ||
| + | <p>The LacI+ strain holds the same construct, but with a LacI gene under the control of Pveg. | ||
| + | The expression of the GFP in this construct depends on the expression level of the tRNA and the basal expression level of the T7amber. </p> | ||
| + | <p> The three strains were cultured overnight, then introduced into a 96-well spectrophotometry plate. The cultures were diluted directly in the wells to an initial OD600 of approximately 0.1 in the presence or absence of IPTG (0.01mM) . The green fluorescence level as well as the OD600 were measured every 5min for 3hrs at 37°C. The 30 minutes delay of manipulation time were taken into consideration in the treatment of the data. </p> | ||
| + | </html> | ||
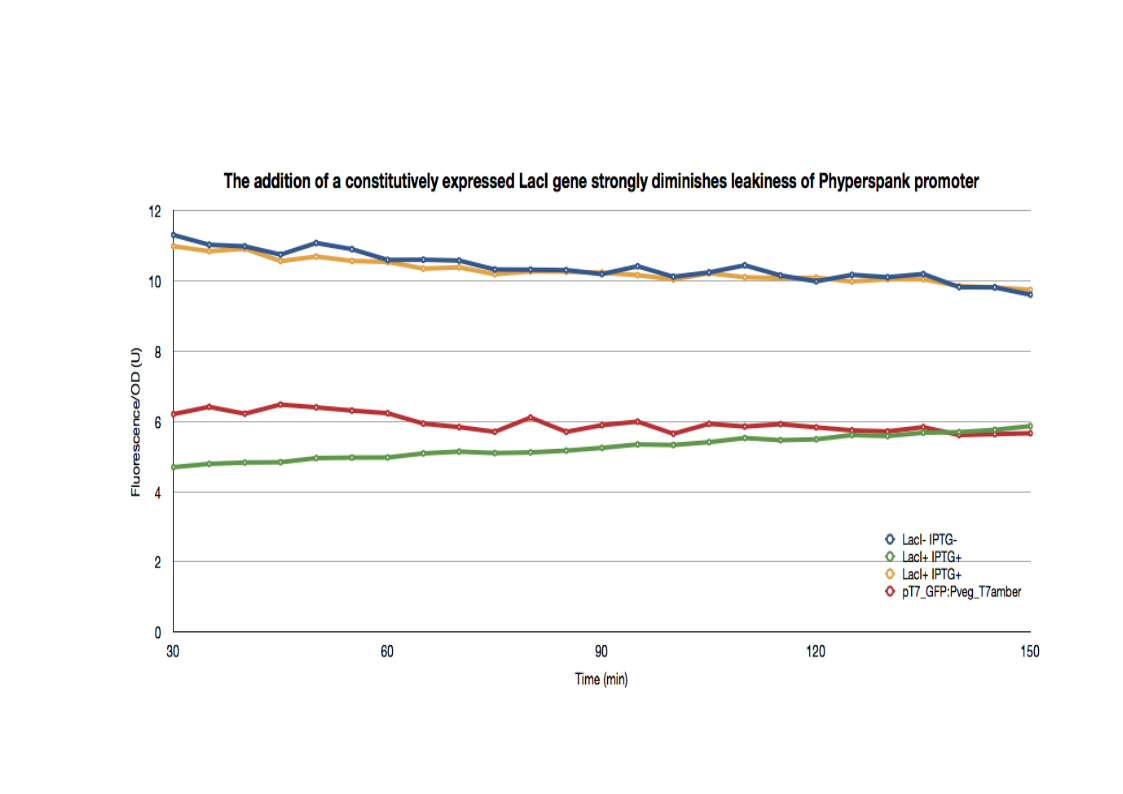
| + | [[File:LacItRNAgraph1.jpg|700px|thumb|center]] | ||
| - | + | <p>The LacI- strains have a basal GFP expression level that is higher than the LacI+ cells. In fact, the GFP expression level in the LacI+ cells are identical to the pT7_GFP:Pveg_T7amber cells. When the same cells are then induced with IPTG, the GFP expression level joins that of the LacI- strains. The | |
| + | addition of a LacI gene completely erases the leakiness of the Phyperspank promoter activity.</p> | ||
<html> | <html> | ||
Latest revision as of 03:45, 29 October 2011

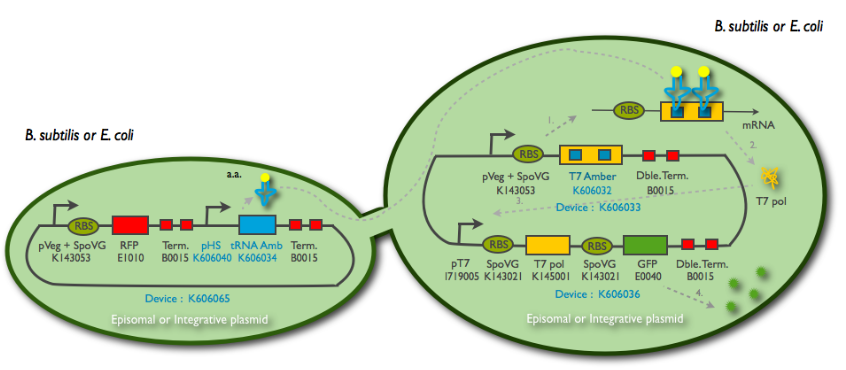
pHyperSpank characterisation
The Hyperspank promoter was designed to be compatible for B. subtilis. It is shorter thant the classic one, but is also recognised by E. coli polymerases.
We cloned it first behind the amber tRNA that would induce a GFP amber on an other plasmid. Then to characterise it we cloned it behind an RFP, into TURBO cells that are LacIq to allow the promoter to be the less leacky possible.
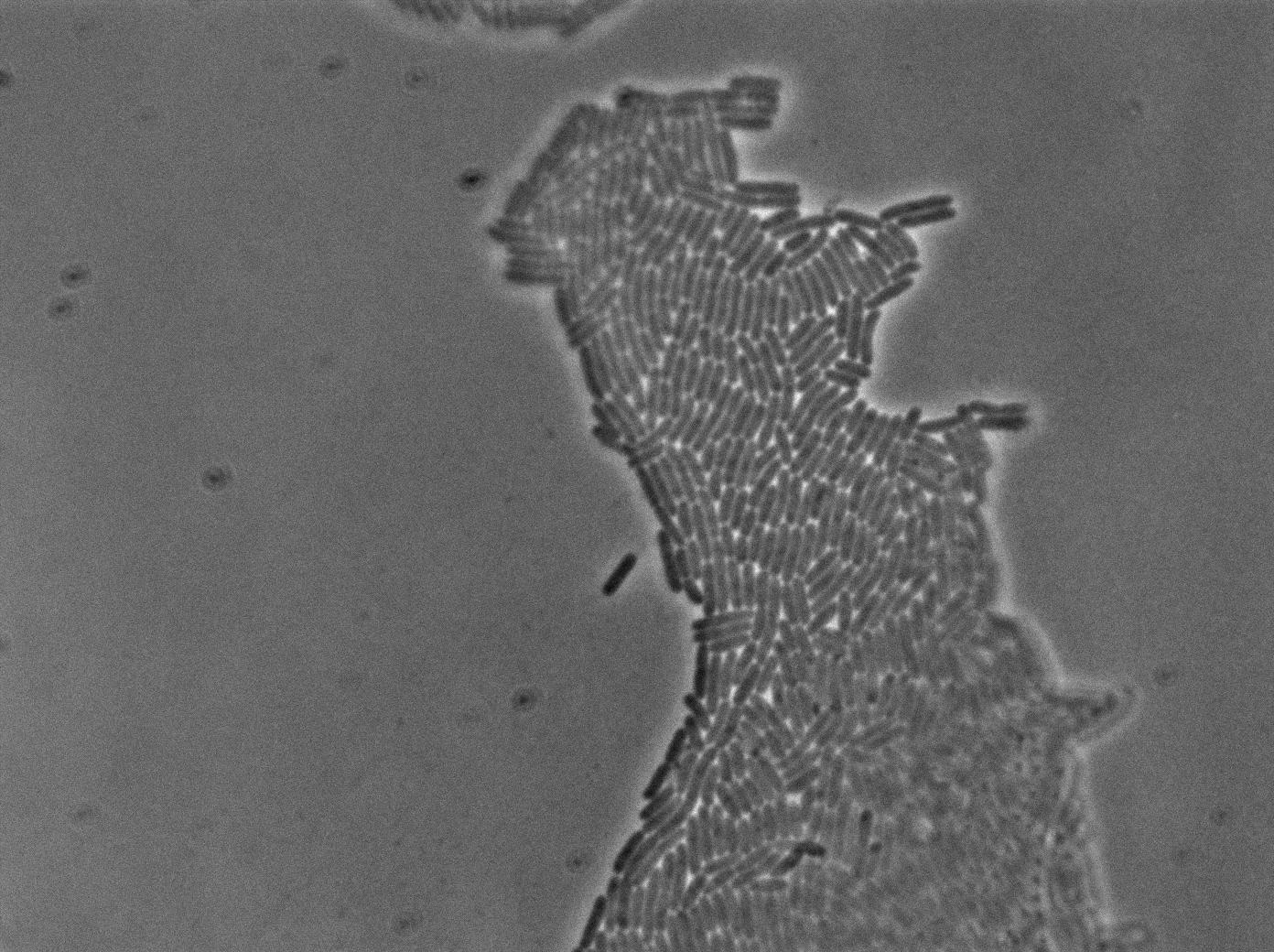
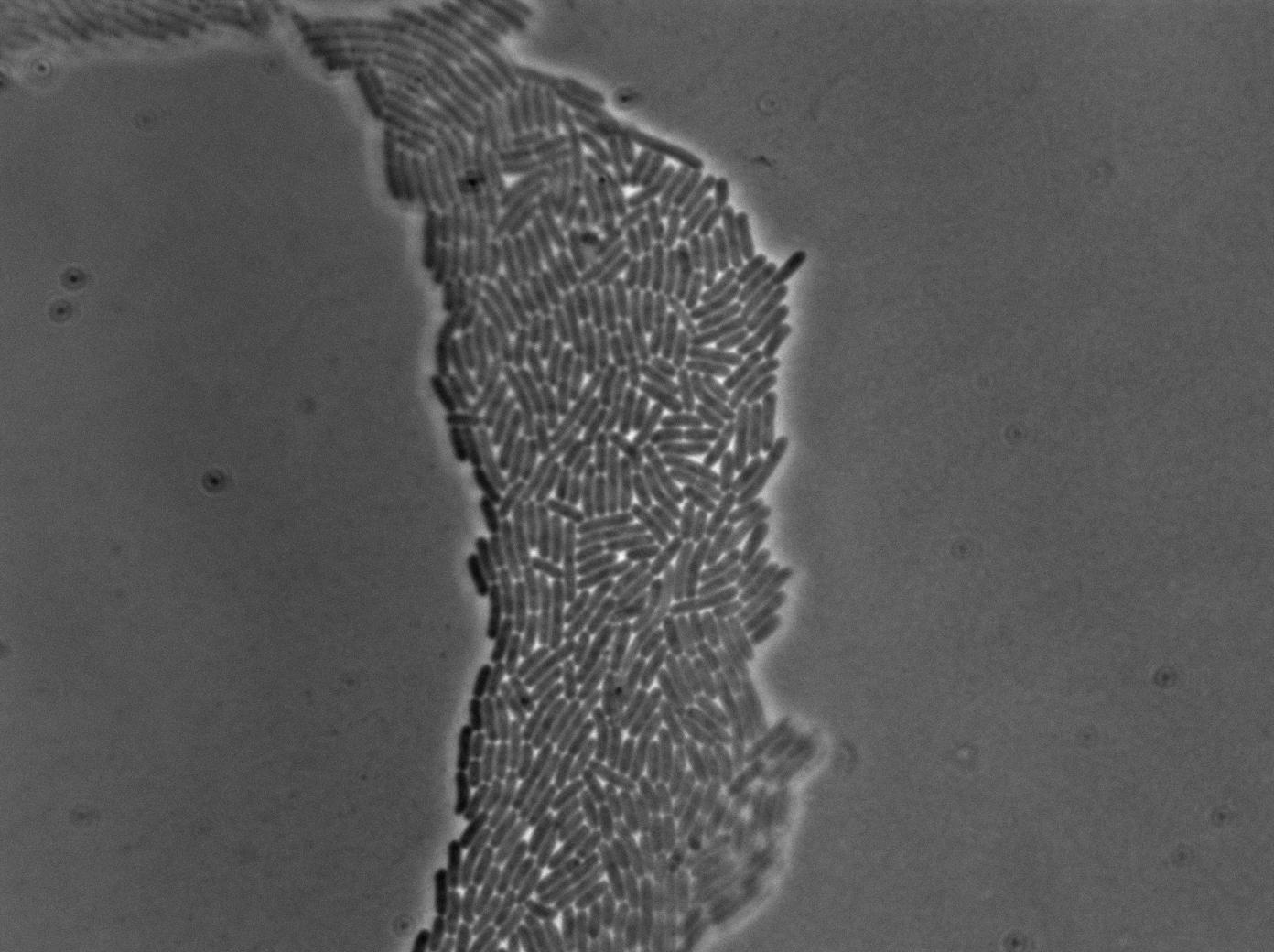
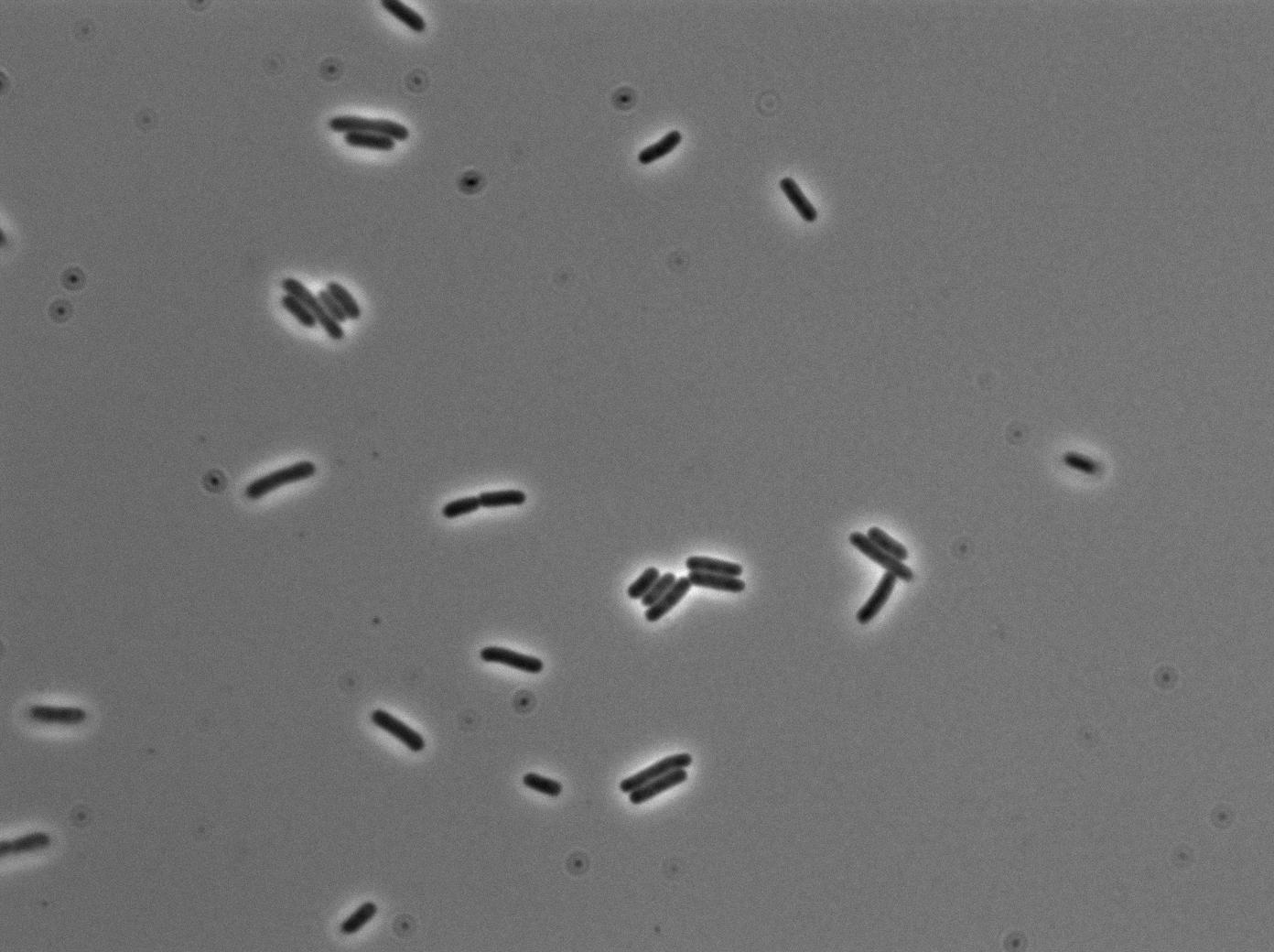
The pictures below are the results of an IPTG induction.
Results
The new biobrick pHS (K606040) was cloned in front of an RFP terminator. The fluorescence was seen under the microscope with strains induced and not induced but IPTG
| E. coli containing Hyperspank +RBS+RFP whithout IPTG at 37°C | |
| E. coli containing Hyperspank +RBS+RFP with IPTG at 37°C | |
The absolute value of the net fluorescence over OD for induced and non indeuced cells was measured in the TECAN i-control, and plotted below.
LacI reduces basal expression of IPTG inducible promoter Phyperspank in E.coli (LacIq+).
Constructs from the tRNA amber system were used for these experiments.
The control strain LacI- holds the construct: pT7_RBS_GFP_tt:Pveg_RBS_T7amber_tt:Phyperspank_tRNA_tt
The control strain pT7_GFP:Pveg_T7amber holds the same construct without the tRNA amber. The GFP expression in this strain solely depends on the basal expression level of the T7amber.
The LacI+ strain holds the same construct, but with a LacI gene under the control of Pveg. The expression of the GFP in this construct depends on the expression level of the tRNA and the basal expression level of the T7amber.
The three strains were cultured overnight, then introduced into a 96-well spectrophotometry plate. The cultures were diluted directly in the wells to an initial OD600 of approximately 0.1 in the presence or absence of IPTG (0.01mM) . The green fluorescence level as well as the OD600 were measured every 5min for 3hrs at 37°C. The 30 minutes delay of manipulation time were taken into consideration in the treatment of the data.
The LacI- strains have a basal GFP expression level that is higher than the LacI+ cells. In fact, the GFP expression level in the LacI+ cells are identical to the pT7_GFP:Pveg_T7amber cells. When the same cells are then induced with IPTG, the GFP expression level joins that of the LacI- strains. The addition of a LacI gene completely erases the leakiness of the Phyperspank promoter activity.
 "
"