Team:Harvard/Technology/One-Hybrid Selection System
From 2011.igem.org
| (6 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Harvard/Template:PracticeBar2}} | {{:Team:Harvard/Template:PracticeBar2}} | ||
| - | + | <div class="whitebox2"> | |
| + | [[Team:Harvard/Results/MAGE | MAGE]] | [[Team:Harvard/Lambda_Red | Lambda Red]]| [[Team:Harvard/Technology/One-Hybrid_Selection System | One-Hybrid Selection]] | [[Team:Harvard/Chip_Based_Library | Chip-Based Library]] | [[Team:Harvard/Protocols | Protocols]] | ||
| + | </div> | ||
| + | <!-- | ||
| + | <div style="height: 21px; width: 937px; background-image: url('http://www.dbgoodman.com/igem2011/images/subbar.png');"> | ||
| + | blah | ||
| + | </div> --> | ||
<div class="whitebox"> | <div class="whitebox"> | ||
| + | PAGE IS DEFUNCT DO NOT EDIT | ||
| + | =One-Hybrid Selection System= | ||
| + | We tested two major selection systems for screening zinc finger binding candidates, the Wolfe selection system utilizing a His3/Ura3 paradigm and the TolC selection system utilizing an SDS/colicin paradigm. Both selection systems are genomic in nature and utilize cell death and survival as part of their selection schemes, making selection straightforward and robust. As shown in the diagram, the genes that form the selection system are regulated upstream by a zinc finger binding site domain. The driving principle behind the selection systems is that a zinc finger binding site is linked to transcription of genes required for cell survival. Thus, cells can only survive if they contain a plasmid expressing zinc fingers that bind specifically enough to the target binding site to increase downstream gene expression. In this way, a library of different zinc finger plasmids can be screened for zinc fingers that bind to the target site due to this event being linked to cell survival. | ||
| - | + | ==Wolfe Selection System (Figure 1)== | |
| - | [[File: | + | [[File: HARVOne-hybrid_diagram_with_caption_for_web.jpg|right|400px|Figure 1: Metabolic Selection Scheme]] |
| - | + | ||
| - | + | ||
| + | The Wolfe selection system utilizes a metabolic selection scheme. In this selection strain, the endogenous HisB, pyrF, and rpoZ genes have been knocked out via lambda-red. This eliminates the cell’s ability to synthesize histidine, pyrF, and the omega-subunit of RNA polymerase, respectively. In the place of HisB and pyrF, the homologous genes His3 and Ura3 have been placed on the genome, with transcription regulated by zinc finger arrays binding upstream. The cells undergo selection in minimal media. Thus, without a zinc finger binding event, the cell is unable to synthesize histidine and dies. To select against a leaky promoter that transcribes the genes in the absence of zinc finger binding, 5-FOA can be added to the selection media. If the promoter is leaky, pyrF will be expressed, and the cell will convert 5-FOA to the toxic product 5-fluorouracil, causing cell death. An alternate method of selecting against false positives is to add 5-AT to the selection media. 5-AT is a selective inhibitor of histidine synthase. Addition of 5-AT would thus select against weak zinc finger binders, leaving only the strongest binders with the ability to active transcription to the level that permits cell survival. | ||
| + | |||
| + | ==TolC Selection System== | ||
| + | Although not a metabolic selection system, the TolC system works similarly, linking gene transcription to cell survival. TolC is a gene encoding for a membrane pump protein that can export toxins such as SDS out of the cell. Selection is accomplished by linking TolC expression to upstream zinc finger binding events while simultaneously introducing SDS into the selection media. Non-specific zinc fingers are unable to active TolC expression, and the cells are thus unable to export SDS out of the cell, leading to cell death. Meanwhile, zinc finger binders successfully activate TolC expression, allowing cells to survive in spite of SDS being present in selection media. Negative selection against leaky promoters is accomplished in the TolC system through addition of colicins to the selection media. Colicins are toxins that require the TolC transporter in order to access the cell. As a result, if the promoter activates TolC transcription in the absence of zinc finger binding, colicins will be allowed to enter the cell, leading to cell death. Fine-tuning of the TolC selection system is accomplished by adding varying amounts of SDS to the selection media, with higher concentrations of SDS only allowing the strongest zinc finger binders to survive. | ||
| + | |||
| + | |||
| + | =Fine-tuning the one-hybrid selection system= | ||
| + | |||
| + | [[File: HARVhybrid1.png|left|300px|Figure 1: Characterization of the selection strain]] | ||
| + | |||
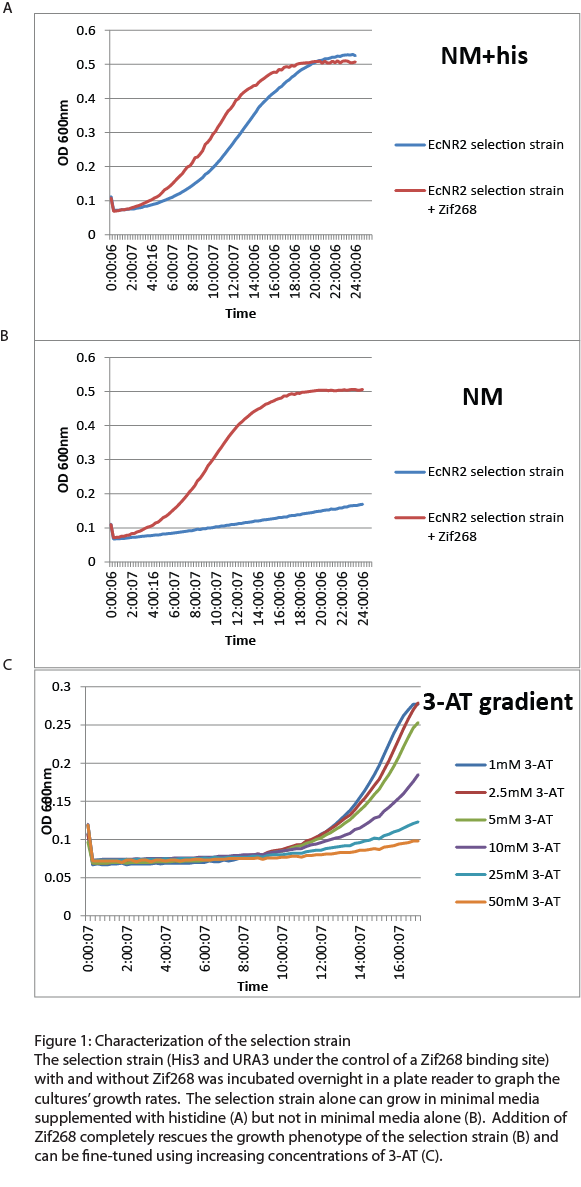
| + | After the one-hybrid His3-URA3 strain was constructed using lambda red and MAGE, we characterized its growth phenotype in preparation for the chip-synthesized zinc fingers. Cultures with the Zif268 binding site either with and without the Zif268 protein were grown overnight in a plate reader to chart their growth under various conditions by measuring the absorbance levels at 600nm. As expected, the strain without Zif268 grew normally in complete media but failed to grow in NM media without histidine (Figure 1A). Zif268 successfully resuced the growth phenotype in NM (Figure 1B) and reached similar saturation levels to cultures grown in NM+histidine. Addition of 5-FOA killed the Zif268 cultures because of their expression of URA3 but did not have a great effect on the selection strain alone, showing that the zinc finger binding site promoter is not inherently leaky. 3-AT, the competitive inhibitor of His3, also fine-tuned selection: Zif268 cultures grew less as 3-AT concentration increased. Overall, the strain showed the proper phenotypes and successfully illustrated selection for zinc finger binding. | ||
| + | [[File: HARVhybrid2.png|right|300px|Figure 2: Simulation of chip conditions]] | ||
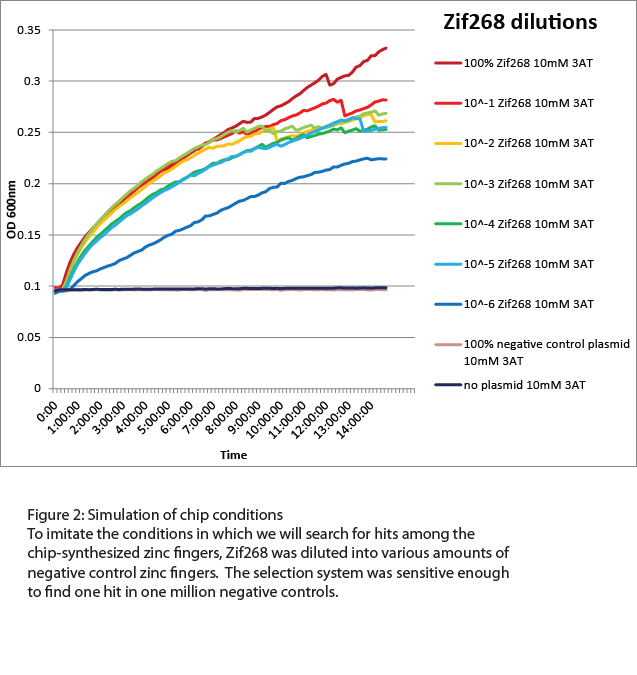
For the zinc fingers synthesized from the chip, the selection strain would need to be able to recognize low numbers of hits among a high level of background. Approximately 9000 different zinc fingers were designed for each binding site, and all 9000 are unlikely to successfully bind to the target, so we tested the sensitivity of the selection strain by diluting Zif268 into zinc finger plasmids that would not bind to the Zif268 site (Figure 2). Overnight growth in a plate reader showed that our selection strain was sensitive enough to detect Zif268 when diluted as low as one to one million in as high a concentration of 3-AT as 10mM. This is more than sufficient to pick out one hit among the 9000 chip-synthesized zinc fingers. | For the zinc fingers synthesized from the chip, the selection strain would need to be able to recognize low numbers of hits among a high level of background. Approximately 9000 different zinc fingers were designed for each binding site, and all 9000 are unlikely to successfully bind to the target, so we tested the sensitivity of the selection strain by diluting Zif268 into zinc finger plasmids that would not bind to the Zif268 site (Figure 2). Overnight growth in a plate reader showed that our selection strain was sensitive enough to detect Zif268 when diluted as low as one to one million in as high a concentration of 3-AT as 10mM. This is more than sufficient to pick out one hit among the 9000 chip-synthesized zinc fingers. | ||
Latest revision as of 16:13, 25 September 2011
MAGE | Lambda Red| One-Hybrid Selection | Chip-Based Library | Protocols
PAGE IS DEFUNCT DO NOT EDIT
Contents |
One-Hybrid Selection System
We tested two major selection systems for screening zinc finger binding candidates, the Wolfe selection system utilizing a His3/Ura3 paradigm and the TolC selection system utilizing an SDS/colicin paradigm. Both selection systems are genomic in nature and utilize cell death and survival as part of their selection schemes, making selection straightforward and robust. As shown in the diagram, the genes that form the selection system are regulated upstream by a zinc finger binding site domain. The driving principle behind the selection systems is that a zinc finger binding site is linked to transcription of genes required for cell survival. Thus, cells can only survive if they contain a plasmid expressing zinc fingers that bind specifically enough to the target binding site to increase downstream gene expression. In this way, a library of different zinc finger plasmids can be screened for zinc fingers that bind to the target site due to this event being linked to cell survival.
Wolfe Selection System (Figure 1)
The Wolfe selection system utilizes a metabolic selection scheme. In this selection strain, the endogenous HisB, pyrF, and rpoZ genes have been knocked out via lambda-red. This eliminates the cell’s ability to synthesize histidine, pyrF, and the omega-subunit of RNA polymerase, respectively. In the place of HisB and pyrF, the homologous genes His3 and Ura3 have been placed on the genome, with transcription regulated by zinc finger arrays binding upstream. The cells undergo selection in minimal media. Thus, without a zinc finger binding event, the cell is unable to synthesize histidine and dies. To select against a leaky promoter that transcribes the genes in the absence of zinc finger binding, 5-FOA can be added to the selection media. If the promoter is leaky, pyrF will be expressed, and the cell will convert 5-FOA to the toxic product 5-fluorouracil, causing cell death. An alternate method of selecting against false positives is to add 5-AT to the selection media. 5-AT is a selective inhibitor of histidine synthase. Addition of 5-AT would thus select against weak zinc finger binders, leaving only the strongest binders with the ability to active transcription to the level that permits cell survival.
TolC Selection System
Although not a metabolic selection system, the TolC system works similarly, linking gene transcription to cell survival. TolC is a gene encoding for a membrane pump protein that can export toxins such as SDS out of the cell. Selection is accomplished by linking TolC expression to upstream zinc finger binding events while simultaneously introducing SDS into the selection media. Non-specific zinc fingers are unable to active TolC expression, and the cells are thus unable to export SDS out of the cell, leading to cell death. Meanwhile, zinc finger binders successfully activate TolC expression, allowing cells to survive in spite of SDS being present in selection media. Negative selection against leaky promoters is accomplished in the TolC system through addition of colicins to the selection media. Colicins are toxins that require the TolC transporter in order to access the cell. As a result, if the promoter activates TolC transcription in the absence of zinc finger binding, colicins will be allowed to enter the cell, leading to cell death. Fine-tuning of the TolC selection system is accomplished by adding varying amounts of SDS to the selection media, with higher concentrations of SDS only allowing the strongest zinc finger binders to survive.
Fine-tuning the one-hybrid selection system
After the one-hybrid His3-URA3 strain was constructed using lambda red and MAGE, we characterized its growth phenotype in preparation for the chip-synthesized zinc fingers. Cultures with the Zif268 binding site either with and without the Zif268 protein were grown overnight in a plate reader to chart their growth under various conditions by measuring the absorbance levels at 600nm. As expected, the strain without Zif268 grew normally in complete media but failed to grow in NM media without histidine (Figure 1A). Zif268 successfully resuced the growth phenotype in NM (Figure 1B) and reached similar saturation levels to cultures grown in NM+histidine. Addition of 5-FOA killed the Zif268 cultures because of their expression of URA3 but did not have a great effect on the selection strain alone, showing that the zinc finger binding site promoter is not inherently leaky. 3-AT, the competitive inhibitor of His3, also fine-tuned selection: Zif268 cultures grew less as 3-AT concentration increased. Overall, the strain showed the proper phenotypes and successfully illustrated selection for zinc finger binding.
For the zinc fingers synthesized from the chip, the selection strain would need to be able to recognize low numbers of hits among a high level of background. Approximately 9000 different zinc fingers were designed for each binding site, and all 9000 are unlikely to successfully bind to the target, so we tested the sensitivity of the selection strain by diluting Zif268 into zinc finger plasmids that would not bind to the Zif268 site (Figure 2). Overnight growth in a plate reader showed that our selection strain was sensitive enough to detect Zif268 when diluted as low as one to one million in as high a concentration of 3-AT as 10mM. This is more than sufficient to pick out one hit among the 9000 chip-synthesized zinc fingers.
We designed an additional one-hybrid selection system utilizing TolC, an SDS pump, under the control of a zinc finger binding site. The strain was successfully made and showed the proper phenotypic rescue when Zif268 was present, but SDS concentrations were not as titratable as 3-AT, and it was not able to recognize hits among high background levels as well as the His3-URA3 system. Due to TolC’s inferior sensitivity, we decided to transform the chip-synthesized zinc fingers only into the His3-URA3 selection strain.
 "
"