Team:DTU-Denmark/Project abstract
From 2011.igem.org
(→Abstract) |
|||
| (2 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{:Team:DTU-Denmark/Templates/Standard_page_begin| | + | {{:Team:DTU-Denmark/Templates/Standard_page_begin|Abstract}} |
== Abstract == | == Abstract == | ||
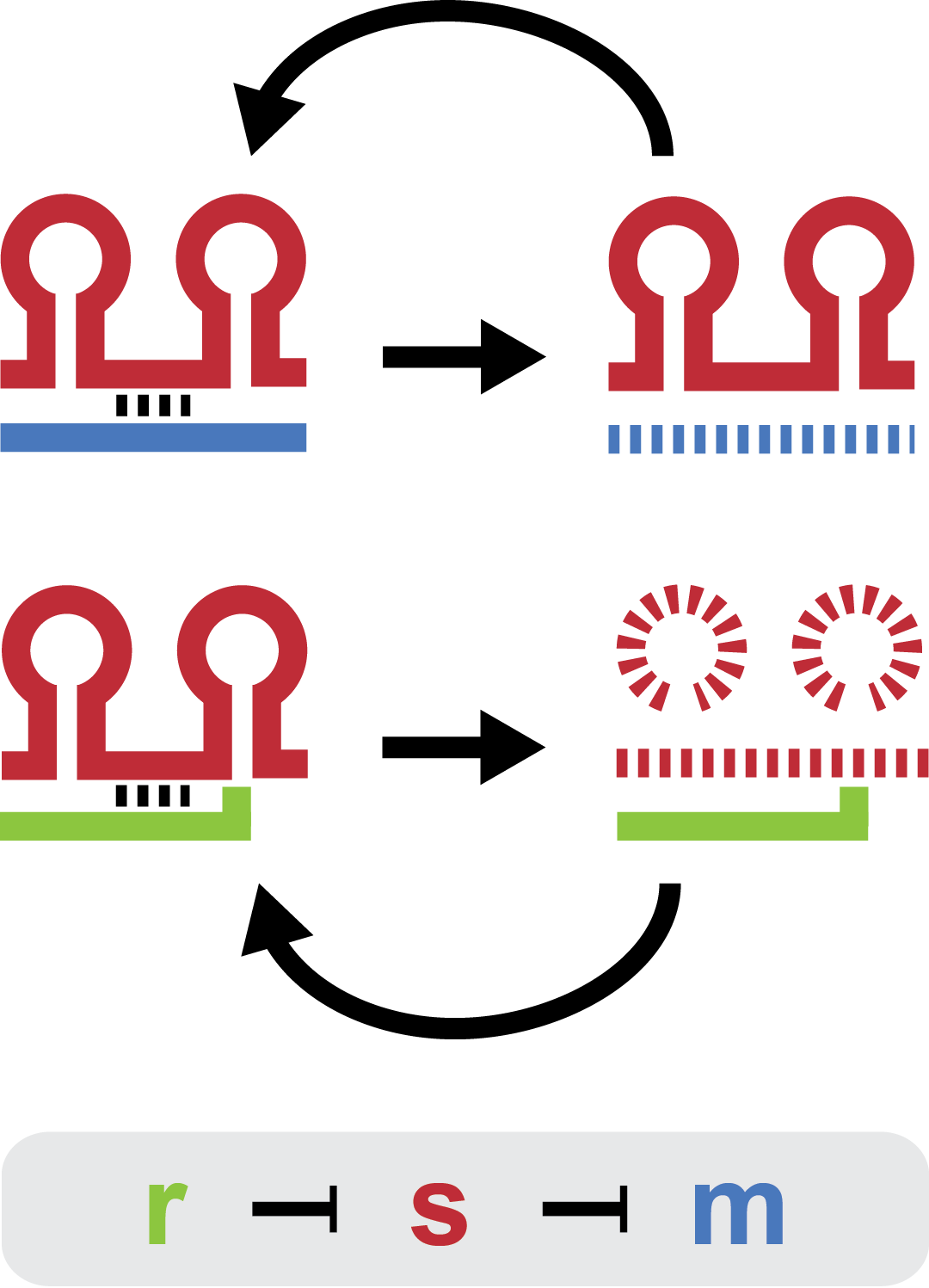
[[File:DTU2011_project_fig1.png|200px|thumb|right|Two-level sRNA regulation. Blue is any target mRNA, red is sRNA and green is trap-RNA.]] | [[File:DTU2011_project_fig1.png|200px|thumb|right|Two-level sRNA regulation. Blue is any target mRNA, red is sRNA and green is trap-RNA.]] | ||
| - | Small regulatory RNA is an active area of research with untapped possibilities for application in biotechnology. Such applications include convenient gene silencing and fine-tuning of synthetic biological circuits, which are currently cumbersome processes restricted to well studied bacteria. We have investigated a novel type of RNA regulation<span class="superscript">[[#References|[1]]]</span><span class="superscript">[[#References|[2]]]</span>, where the inhibition caused by a small regulatory RNA is relieved by another RNA called trap-RNA. The system displays a large dynamic range and can uniquely target and repress any gene of interest providing unprecedented flexibility. We suspect that any level of repression is achievable by simply altering the sequences of the involved RNAs. Multiple such systems can coexist without interfering and are thus compatible with more complex designs. | + | Small regulatory RNA is an active area of research with untapped possibilities for application in biotechnology. Such applications include convenient gene silencing and fine-tuning of synthetic biological circuits, which are currently cumbersome processes restricted to well studied bacteria. We have investigated a novel type of RNA regulation<span class="superscript">[[#References|[1]]]</span><span class="superscript">[[#References|[2]]]</span>, where the inhibition caused by a small regulatory RNA is relieved by another RNA called trap-RNA. The system displays a large dynamic range and can uniquely target and repress any gene of interest providing unprecedented flexibility. We suspect that any level of repression is achievable by simply altering the sequences of the involved RNAs. Multiple such systems can coexist without interfering and are, thus, compatible with more complex designs. |
Latest revision as of 19:45, 21 September 2011
Abstract
Abstract
Small regulatory RNA is an active area of research with untapped possibilities for application in biotechnology. Such applications include convenient gene silencing and fine-tuning of synthetic biological circuits, which are currently cumbersome processes restricted to well studied bacteria. We have investigated a novel type of RNA regulation[1][2], where the inhibition caused by a small regulatory RNA is relieved by another RNA called trap-RNA. The system displays a large dynamic range and can uniquely target and repress any gene of interest providing unprecedented flexibility. We suspect that any level of repression is achievable by simply altering the sequences of the involved RNAs. Multiple such systems can coexist without interfering and are, thus, compatible with more complex designs.
References
[1] Figueroa-Bossi, Nara, Martina Valentini, Laurette Malleret, and Lionello Bossi. “Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target.” Genes & Development 23, no. 17 (2009): 2004 -2015.
[2] Overgaard, Martin, Jesper Johansen, Jakob Møller‐Jensen, and Poul Valentin‐Hansen. “Switching off small RNA regulation with trap‐mRNA.” Molecular Microbiology 73, no. 5 (September 2009): 790-800.
 "
"