Team:EPF-Lausanne/Our Project/T7 promoter variants
From 2011.igem.org
(→DNA Recovery with Lysis) |
|||
| (32 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{:Team:EPF-Lausanne/Templates/ | + | {{:Team:EPF-Lausanne/Templates/T7lysisHeader|title=Intro}} |
| - | + | One major challenge in designing new regulatory parts is to determine which combinations of transcription factors and binding sequences match. | |
| + | From previous research and our own MITOMI experiments, we know which DNA sequences TetR binds to, and which residues of TetR participate in binding, but we do not know how changing these residues will affect either binding affinity or specificity. | ||
| + | Molecular dynamics simulations and other theoretical approaches have not come any closer to answering these questions. | ||
| + | In short, we know too little about protein-DNA interaction to intelligently design transcription factors. | ||
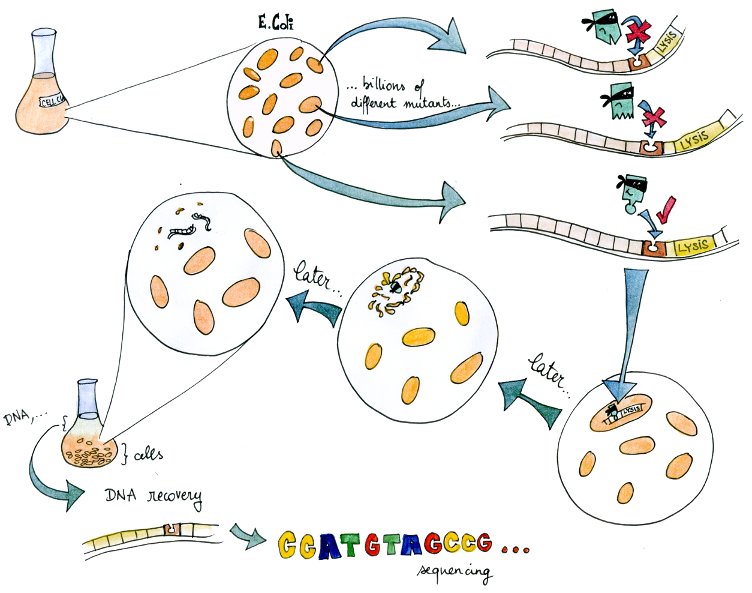
| + | To make up for this lack of knowledge, we present an experimental system to select valid binding pairs from many random tetR and pTet mutants, based on an inducible lysis gene. | ||
| - | + | [[File:EPFL-Solange-Lysis.jpg|700px]] | |
| - | + | The system – in a way a "survival of the weakest" – is related to directed evolution. | |
| + | A lysis system based on the K112808 lysis device is indirectly activated by tetR. | ||
| + | Therefore, if in a given cell the tetR variant present can bind to the tetR promoter, the cell lyses and releases its DNA into the culture media. | ||
| + | From there, DNA can be recovered and amplified, tranformed, or directly sequenced. | ||
| + | By design, this DNA codes for a combination of TF and promoter with high mutual affinity, and therefore almost directly yields a valid regulatory part. | ||
| + | In this light, it is a useful component of our transcription factor development pipeline. | ||
| - | + | This is a direct and practical way of solving the problem of selecting high affinity pairs among the millions of possible combinations of transcription factors and promoters. | |
| + | It can be seen as a form of DNA-based information processing, and is therefore also a neat example of a problem more efficiently solved by non-conventional computation. | ||
| - | + | To develop the system, we began with a [[Team:EPF-Lausanne/Our_Project/T7_promoter_variants/lysis|simple experiment]] to check that a lysis cassette in a plasmid could lyse with greater efficiency as a function of IPTG concentration. The next step towards our goal was to demonstrate that [[Team:EPF-Lausanne/Our_Project/T7_promoter_variants/recovery|plasmid DNA can be adequately recovered]] and repackaged (PCR amplified, transformed into a different strain, etc...) as a result of lysing. In a [[Team:EPF-Lausanne/Our_Project/T7_promoter_variants/selection|more elaborate experiment]], we were able to show that not only did the lysing efficiently release plasmids from the cells, but that it could be made to do so selectively in a large culture containing a variety of strains. Finally, cognizant of the fact that a good lysis selection method ought to be flexible with regards to the larger reporter system, we manufactured [[Team:EPF-Lausanne/Our_Project/T7_promoter_variants/t7prom|twelve different T7 promoter variants]] that exhibit a wide range of strengths and induction efficiencies. The latter will play a crucial role in being able to accomodate the activation time-scales of fragile and complex selection systems. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | [[ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | In | + | |
| - | + | ||
| - | [[ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | [[ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | All the components of the selection machine have been separately tested experimentally and found to work. Therefore we are convinced the system can work, but it would still require a full-circle experiment to demonstrate its usefulness. | ||
{{:Team:EPF-Lausanne/Templates/Footer}} | {{:Team:EPF-Lausanne/Templates/Footer}} | ||
Latest revision as of 03:03, 22 September 2011
Lysis Selection System
Lysis selection system Main | Lysis Characterization | DNA Recovery | DNA Selection | T7 Promoter VariantsOne major challenge in designing new regulatory parts is to determine which combinations of transcription factors and binding sequences match. From previous research and our own MITOMI experiments, we know which DNA sequences TetR binds to, and which residues of TetR participate in binding, but we do not know how changing these residues will affect either binding affinity or specificity. Molecular dynamics simulations and other theoretical approaches have not come any closer to answering these questions. In short, we know too little about protein-DNA interaction to intelligently design transcription factors. To make up for this lack of knowledge, we present an experimental system to select valid binding pairs from many random tetR and pTet mutants, based on an inducible lysis gene.
The system – in a way a "survival of the weakest" – is related to directed evolution. A lysis system based on the K112808 lysis device is indirectly activated by tetR. Therefore, if in a given cell the tetR variant present can bind to the tetR promoter, the cell lyses and releases its DNA into the culture media. From there, DNA can be recovered and amplified, tranformed, or directly sequenced. By design, this DNA codes for a combination of TF and promoter with high mutual affinity, and therefore almost directly yields a valid regulatory part. In this light, it is a useful component of our transcription factor development pipeline.
This is a direct and practical way of solving the problem of selecting high affinity pairs among the millions of possible combinations of transcription factors and promoters. It can be seen as a form of DNA-based information processing, and is therefore also a neat example of a problem more efficiently solved by non-conventional computation.
To develop the system, we began with a simple experiment to check that a lysis cassette in a plasmid could lyse with greater efficiency as a function of IPTG concentration. The next step towards our goal was to demonstrate that plasmid DNA can be adequately recovered and repackaged (PCR amplified, transformed into a different strain, etc...) as a result of lysing. In a more elaborate experiment, we were able to show that not only did the lysing efficiently release plasmids from the cells, but that it could be made to do so selectively in a large culture containing a variety of strains. Finally, cognizant of the fact that a good lysis selection method ought to be flexible with regards to the larger reporter system, we manufactured twelve different T7 promoter variants that exhibit a wide range of strengths and induction efficiencies. The latter will play a crucial role in being able to accomodate the activation time-scales of fragile and complex selection systems.
All the components of the selection machine have been separately tested experimentally and found to work. Therefore we are convinced the system can work, but it would still require a full-circle experiment to demonstrate its usefulness.
 "
"