Team:Washington/Alkanes/Future/LuxCDE
From 2011.igem.org
(→Gibson Assembly) |
|||
| (21 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
__NOTOC__ | __NOTOC__ | ||
| - | =Alternative Routes for Fatty Aldehyde Production= | + | ='''<center>Alternative Routes for Fatty Aldehyde Production'''= |
==Background== | ==Background== | ||
| Line 9: | Line 9: | ||
[[File:Washington_luxcde_diagram.png|right|300px|thumb|A diagram of the LuxCDE pathway, producing tetradecanal, the aldehyde substrate for use with aldehyde decarbonalase (ADC)]] | [[File:Washington_luxcde_diagram.png|right|300px|thumb|A diagram of the LuxCDE pathway, producing tetradecanal, the aldehyde substrate for use with aldehyde decarbonalase (ADC)]] | ||
| - | LuxABCDE is an enzyme complex submitted to the iGem registry by the 2010 Cambridge team. Within this complex, LuxAB produces luciferase (used by | + | LuxABCDE is an enzyme complex submitted to the iGem registry by the 2010 Cambridge team. Within this complex, LuxAB produces luciferase (used by [https://2010.igem.org/Team:Cambridge/Bioluminescence/Bacterial_Luciferases Cambridge's] EGlowi) using 14-carbon aldehydes (tetradecanal) produced by LuxCDE. Since the aldehyde decarbonylase (ADC) in our project uses aldehydes to synthesize alkanes, we plan to use the LuxCDE complex in conjunction with ADC to produce 13-carbon alkanes (Tridecane). |
| - | + | <br> | |
==Methods== | ==Methods== | ||
===PCR and Standard Biobrick Assembly=== | ===PCR and Standard Biobrick Assembly=== | ||
| - | This method revolved around designing oligos to PCR amplify LuxCD and LuxE off of the LuxBrick; LuxE was not directly adjacent to LuxCD in the Luxbrick, and therefore had to be amplified out in two separate reactions. Homology on the ends of the two fragments would | + | This method revolved around designing oligos to PCR amplify LuxCD and LuxE off of the LuxBrick; LuxE was not directly adjacent to LuxCD in the Luxbrick, and therefore had to be amplified out in two separate reactions. Homology on the ends of the two fragments would allow us to obtain a LuxCDE using stitching PCR, then ligate this fragment and ADC into psb1C3. We would then assay for alkanes. |
| + | |||
| + | <br> | ||
===Gibson Assembly=== | ===Gibson Assembly=== | ||
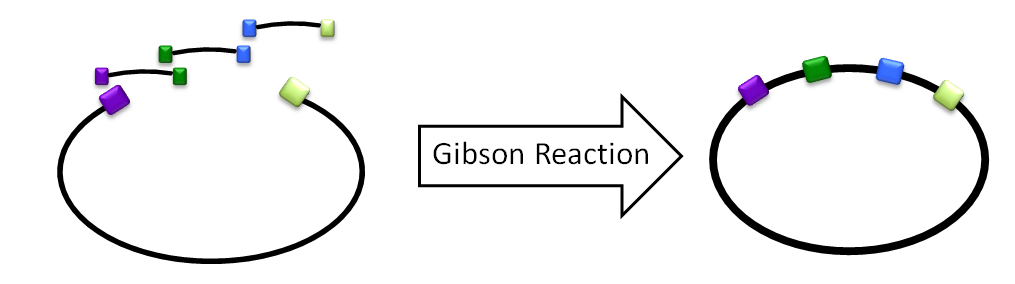
| - | [[File:Washington2011 GibsonReaction.png|right|300px|thumb|A diagram of the | + | [[File:Washington2011 GibsonReaction.png|right|300px|thumb|A basic diagram of a Gibson reaction. All components of the desired vector are left to ligate simultaneously, then transformed into cells.]] |
| - | We would design oligos to amplify C, D, and E off of the LuxBrick. | + | We would design oligos with homology to amplify C, D, and E off of the LuxBrick. The homology would allow the enzymes in the Gibson reaction to chew back the homologous ends and ligate them togather. A 4-piece Gibson assembly was done on C, D, E, and the plasmid backbone with along with aldehyde decarbonylase and the product was then transformed into cells. Another reaction we tried was to amplify everything on the LuxBrick except luxA and luxB. In this reaction we then switched the two genes with ADC to create a product with luxC, luxD, luxE, luxG and ADC. |
| + | |||
| + | <br> | ||
===Gene Synthesis=== | ===Gene Synthesis=== | ||
| - | Instead of using the LuxBrick to obtain C, D, and E, | + | Instead of using the LuxBrick to obtain C, D, and E, C, D, and E was re-optimized to have cloning-friendly GC content. Primers were designed to synthesize the genes separately each in their own PCR reaction. Another PCR was used to amplify the respective pieces, and each piece was ligated into psb1C3. Once each gene was in psb1C3, it was re-amplified out and ligated into a vector containing ADC. This is done step wise until the full construct is obtained. |
| + | |||
| + | <br> | ||
==Results== | ==Results== | ||
| - | The PCR and Gibson methods failed at multiple steps for uncertain reasons. Since we do not have our LuxCDE construct, no method or assay for testing the hypothetical construct was developed as of this time. We hypothesize that LuxCD's abnormally high AT concentration may have caused misannealing for primers, causing primer-dimer errors and other problems, but no concrete evidence exists for this as of yet. For gene assembly, we have lowered the AT concentration of C, D, and E to more optimal levels for PCR. To be precise, the C, D, and E had an AT content of 66.3, 65.0, and 66.9 percent, which was reduced to 50.2, 50.1, and 51.6 percent respectively. | + | The PCR and Gibson methods failed at multiple steps for uncertain reasons. Since we do not have our LuxCDE construct, no method or assay for testing the hypothetical construct was developed as of this time. We hypothesize that LuxCD's abnormally high AT concentration may have caused misannealing for primers, causing primer-dimer errors and other problems, but no concrete evidence exists for this as of yet. For gene assembly, we have lowered the AT concentration of C, D, and E to more optimal levels for PCR. To be precise, the C, D, and E had an AT content of 66.3, 65.0, and 66.9 percent, which was reduced to 50.2, 50.1, and 51.6 percent respectively. Re-optimization likely helped cleanly amplifying out the genes. |
| - | + | <br> | |
==Current Status== | ==Current Status== | ||
| - | Attempts to clone LuxCDE using standard PCR/Biobrick Assembly and Gibson Assembly have been abandoned. Gene assembly was successful and each of the three luxCDE genes have been sequenced, biobricked and submitted. | + | Attempts to clone LuxCDE using standard PCR/Biobrick Assembly and Gibson Assembly have been abandoned. Gene assembly was successful and each of the three luxCDE genes have been sequenced, biobricked and submitted. We are currently working on assembling the full CDE + ADC construct. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<html> | <html> | ||
| - | <img src=https://static.igem.org/mediawiki/2011/ | + | <img src=https://static.igem.org/mediawiki/2011/e/eb/Washington_cde_construct.png width="750" height="353" usemap="#washington cde construct.png"/> |
| - | <map name="washington | + | <map name="washington cde construct.png"> |
| + | <area shape="rect" coords="143,99,294,250" title="Part:BBa_K590031" href="http://partsregistry.org/Part:BBa_K590031"/> | ||
| + | <area shape="rect" coords="48,107,143,201" title="Part:BBa_J23100" href="http://partsregistry.org/Part:BBa_J23100"/> | ||
| + | <area shape="rect" coords="294,99,446,250" title="Part:BBa_K590059" href="http://partsregistry.org/Part:BBa_K590059"/> | ||
| + | <area shape="rect" coords="446,99,598,250" title="Part:BBa_K590060" href="http://partsregistry.org/Part:BBa_K590060"/> | ||
| + | <area shape="rect" coords="598,99,750,250" title="Part:BBa_K590061" href="http://partsregistry.org/Part:BBa_K590061"/> | ||
</map> | </map> | ||
</html> | </html> | ||
| + | |||
| + | ==Parts Submitted== | ||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590059 LuxC], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590060 LuxD], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590061 LuxE] | ||
Latest revision as of 02:22, 29 September 2011
Alternative Routes for Fatty Aldehyde Production
Background
LuxABCDE is an enzyme complex submitted to the iGem registry by the 2010 Cambridge team. Within this complex, LuxAB produces luciferase (used by Cambridge's EGlowi) using 14-carbon aldehydes (tetradecanal) produced by LuxCDE. Since the aldehyde decarbonylase (ADC) in our project uses aldehydes to synthesize alkanes, we plan to use the LuxCDE complex in conjunction with ADC to produce 13-carbon alkanes (Tridecane).
Methods
PCR and Standard Biobrick Assembly
This method revolved around designing oligos to PCR amplify LuxCD and LuxE off of the LuxBrick; LuxE was not directly adjacent to LuxCD in the Luxbrick, and therefore had to be amplified out in two separate reactions. Homology on the ends of the two fragments would allow us to obtain a LuxCDE using stitching PCR, then ligate this fragment and ADC into psb1C3. We would then assay for alkanes.
Gibson Assembly
We would design oligos with homology to amplify C, D, and E off of the LuxBrick. The homology would allow the enzymes in the Gibson reaction to chew back the homologous ends and ligate them togather. A 4-piece Gibson assembly was done on C, D, E, and the plasmid backbone with along with aldehyde decarbonylase and the product was then transformed into cells. Another reaction we tried was to amplify everything on the LuxBrick except luxA and luxB. In this reaction we then switched the two genes with ADC to create a product with luxC, luxD, luxE, luxG and ADC.
Gene Synthesis
Instead of using the LuxBrick to obtain C, D, and E, C, D, and E was re-optimized to have cloning-friendly GC content. Primers were designed to synthesize the genes separately each in their own PCR reaction. Another PCR was used to amplify the respective pieces, and each piece was ligated into psb1C3. Once each gene was in psb1C3, it was re-amplified out and ligated into a vector containing ADC. This is done step wise until the full construct is obtained.
Results
The PCR and Gibson methods failed at multiple steps for uncertain reasons. Since we do not have our LuxCDE construct, no method or assay for testing the hypothetical construct was developed as of this time. We hypothesize that LuxCD's abnormally high AT concentration may have caused misannealing for primers, causing primer-dimer errors and other problems, but no concrete evidence exists for this as of yet. For gene assembly, we have lowered the AT concentration of C, D, and E to more optimal levels for PCR. To be precise, the C, D, and E had an AT content of 66.3, 65.0, and 66.9 percent, which was reduced to 50.2, 50.1, and 51.6 percent respectively. Re-optimization likely helped cleanly amplifying out the genes.
Current Status
Attempts to clone LuxCDE using standard PCR/Biobrick Assembly and Gibson Assembly have been abandoned. Gene assembly was successful and each of the three luxCDE genes have been sequenced, biobricked and submitted. We are currently working on assembling the full CDE + ADC construct.

 "
"