Team:Washington/Alkanes/Future/DecarbDesign
From 2011.igem.org
(→Current Status) |
|||
| (64 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:Team:Washington/Templates/Top}} | {{Template:Team:Washington/Templates/Top}} | ||
| + | __NOTOC__ | ||
| - | <big> | + | <center><big><big><big><big>Decarbonylase Redesign</big></big></big></big></center> |
='''Background'''= | ='''Background'''= | ||
| - | <nowiki> | + | :<nowiki>Alkane biosynthesis, as it is, is mainly limited by ADC’s activity. In an effort to diversify alkane production, we strived to improve ADC’s activity on shorter fatty aldehydes.</nowiki> |
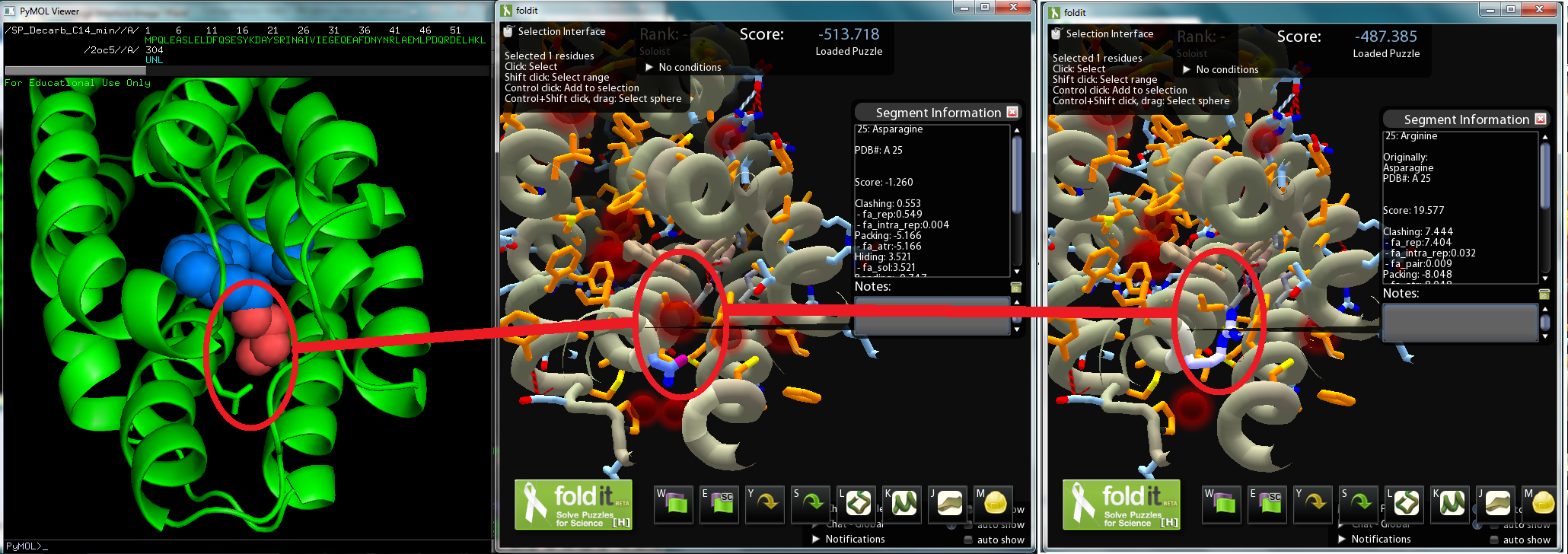
| - | + | :A crystal structure derived model of ADC was put on a Pymol file and reveals a octadecanoic acid stuck in the protein. The octadecanoic acid was modified to a tetradecanal, and then the Pymol file was converted to a Fold-it file for human interaction. On Fold-it, we can model mutations of amino acids in the enzyme and Fold-it computes a relative value representing the enzyme’s stability. As we wanted to maintain the aldehyde decarbonylation, we avoided changing amino acids near the catalytic iron center that binds to the aldehyde group. | |
| - | + | :The most promising mutation sites seem to be amino acids 21 to 25 and 67 to 71, sections of two α-helices adjacent to the tetradecanal's carbon end; by mutating those amino acids to ones with larger side-chains, tetradecanal might fit better in ADC. | |
| - | + | ||
| - | + | [[File:Washington_2011_Redesign_Interface_Mutation.png|center|800x500px|thumb|Screenshot of Pymol model(left), Fold-It model of ADC (center), Fold it model of ADC mutant (right)]] | |
='''Methods'''= | ='''Methods'''= | ||
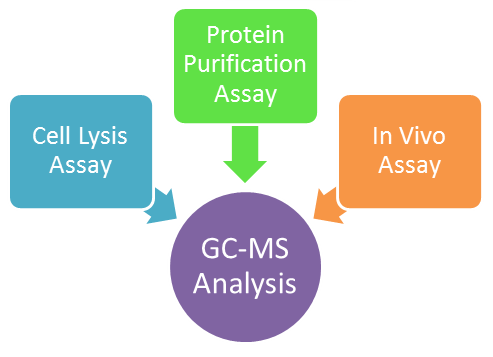
[[File:Washington_redesign_JHA.png|right|300x300px|thumb|Three types of assays were developed and analyzed on Gas chromatography–mass spectrometry (GC-MS)]] | [[File:Washington_redesign_JHA.png|right|300x300px|thumb|Three types of assays were developed and analyzed on Gas chromatography–mass spectrometry (GC-MS)]] | ||
| - | + | After using Fold-It, a computational protein design program, to model several ADC mutants, the ADC mutants were made using Kunkel Mutagenesis. The sequences of these mutants were later verified to carry the correct mutations. After creating several ADC mutants, three different types of assays were developed and the mutants were later tested and analyzed using Gas chromatography–mass spectrometry (GC-MS) to see if our mutants can produce more C13 alkanes. Three different assays were set up: cell lysis assay, protein purification assay, and in vivo assay. | |
| + | |||
==In Vitro Assays== | ==In Vitro Assays== | ||
| + | In these assays, ADC was cloned into a vector with an inducible promoter and Kanamycin resistance. | ||
*<big> Cell Lysis Assay </big> | *<big> Cell Lysis Assay </big> | ||
| - | + | One aldehyde decarbonylase (ADC) colony was picked and grown in the 37°C shaker until OD600 reached the value of 0.8. Then, the culture was induced with IPTG and cell pellet was collected by spinning down after three more hours of shaking. This cell pellet was resuspended in a solution that contains sodium phosphate buffer, protease inhibitor, DNAses and lysozyme. The resuspended cells were lysed with 10% triton then centrifuged. The supernatant was transferred to a glass vial and used to set up in vitro reactions. In vitro reactions were set up by adding certain volume of cells, feredoxin, feredoxin reductase, NADPH and aldehyde. | |
| + | However, this assay did not yield consistent alkane production every time even though we kept variables very minimal. So our team decided to move on and develop a different assay, protein purification assay. | ||
*<big> Protein Purification Assay </big> | *<big> Protein Purification Assay </big> | ||
| - | We grow up cells to express 10 times more ADC proteins | + | We decided to increase the concentration of ADC and decrease the number of contaminants in the assay. We grow up cells to express 10 times more ADC proteins and then we purify the proteins by lysing the cells and binding the target ADC proteins to the BioRad columns and then eluting them out, resulting in pure ADC proteins. Then, we setted up in vitro reactions by adding purified ADC mutant protein, standard buffer(Hepes at pH8, NaCl, glycerol, TCEP and water) and feredoxin, feredoxin reductase, NADPH, and aldehyde into an eppendorf. After certain time period has passed, it was quenched in ethyl acetate and the top organic layer that results from quencing was extracted to an insert in a GC-MS vial. This was later analyzed in the GC-MS. |
==In Vivo Assay== | ==In Vivo Assay== | ||
| - | + | We took the base construct RED-PSB1C3 (Acyl-ACP Reductase in PSB1C3 high constitutive vector) and made the 1C3-const-RED-rbs-mutant decarb construct (1C3 construct with RED, ribosomal binding site, and the mutant decarbonylase genes made with Kunkel (site directed) mutagenesis. Then we transformed the competent cells (BL-21) to take up construct plasmids with our mutated ADC genes. We then grow the transformed competent cells in TB with chloramphenicol and then plate it on TB-chloramphenicol agar plates. Individual colonies were taken to inoculate water, and the colony waters were taken to run colony PCRs to confirm the uptake of the correct plasmid constructs that are with mutated ADC genes. Once verified, the corresponding colony waters of the successful colonies were grown in TB-chloramphenicol media overnight. The media are then miniprepped to obtain DNA of the original plasmid construct taken up by BL-21. The miniprepped plasmid construct is then transformed into another form of the competent cells (MG1655) and then plated on TB-chloramphenicol agar to have samples of different genetic variety of samples. Six isolated colonies are picked on each plate (to average out genetic differences) to be grown in special alkane production media M9 for 48 hours. After that, alkane is extracted with ethyl acetate and the samples are then analyzed using GC-MS. | |
| - | + | ='''Current Status & Results'''= | |
| - | We made | + | We have made and tested 5 mutant Aldehyde Decarbonylases. However, due to time constraints and insufficient protein storage conditions, we lack usable data from the purified protein assays. It's possible that ADC loses effectiveness quickly even when stored. |
| + | From our experience, if one's sole purpose is to test mutants, the best thing to do is use a purified protein assay with C14 aldehydes to test improved activity, then characterize the wild type ADC and the promising mutants with C16 aldehyde assays and C18 aldehyde assays. An in vivo assay may work if LuxCDE constructs are made, as LuxCDE reportedly produces a C14 aldehyde. | ||
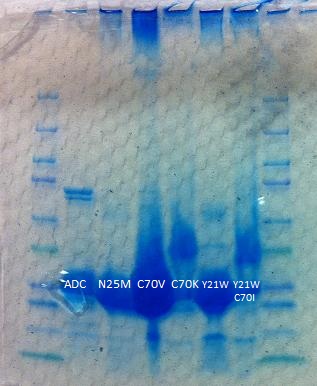
| - | + | [[File:Washington_2011_ADCredesign_proteingel.jpg|left|290px|thumb|Above is the protein gel ran after purifying aldehyde decarbonylase (ADC). Because there isn't a continuous band of proteins, it indicates that we attained a purer sample of ADC. The stray bands are probably impurities.]] | |
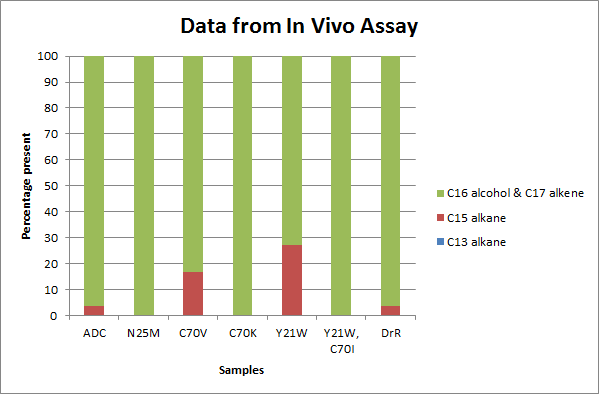
| - | + | [[File:Washington_Redesign_Data1.png|right|900px|thumb|While the mutations definitely showed altered substrate specificity due to different ratios of activity, due to the construct's low or even lacking production on C13 alkanes, the in vivo assay does not say for sure if the specificity was skewed toward shorter or longer aldehyde decarbonylation.]] | |
| - | + | ||
| - | + | ||
| - | + | ||
='''Parts Submitted'''= | ='''Parts Submitted'''= | ||
| - | * | + | We have submitted our five Aldehyde Decarbonylase mutants. |
| - | ** | + | *[http://partsregistry.org/wiki/index.php?title=Part:BBa_K590037 Aldehyde Decarbonylase_N25M] |
| - | ** | + | *[http://partsregistry.org/wiki/index.php?title=Part:BBa_K590038 Aldehyde Decarbonylase_C70V] |
| - | ** | + | *[http://partsregistry.org/wiki/index.php?title=Part:BBa_K590039 Aldehyde Decarbonylase_C70K] |
| - | * | + | *[http://partsregistry.org/wiki/index.php?title=Part:BBa_K590040 Aldehyde Decarbonylase_Y21W] |
| - | * | + | *[http://partsregistry.org/wiki/index.php?title=Part:BBa_K590040 Aldehyde Decarbonylase_Y21W, C70I] |
| - | ** | + | |
| + | Below are the submitted parts that co-express Aldehyde Decarbonylase(normal or mutant) and Acyl-ACP Reductase, the two genes responsible for alkane production. | ||
| + | *[http://partsregistry.org/wiki/index.php?title=Part:BBa_K590053 RrD_PSB1C3-High constitutive] | ||
| + | *[http://partsregistry.org/wiki/index.php?title=Part:BBa_K590054 RrD_N25M-PSB1C3-High constitutive] | ||
| + | *[http://partsregistry.org/wiki/index.php?title=Part:BBa_K590055 RrD_C70V-PSB1C3-High constitutive] | ||
| + | *[http://partsregistry.org/wiki/index.php?title=Part:BBa_K590056 RrD_C70K-PSB1C3-High constitutive] | ||
| + | *[http://partsregistry.org/wiki/index.php?title=Part:BBa_K590057 RrD_Y21W-PSB1C3-High constitutive] | ||
| + | *[http://partsregistry.org/wiki/index.php?title=Part:BBa_K590058 RrD_Y21W, C70I-PSB1C3-High constitutive] | ||
Latest revision as of 19:20, 26 September 2011
Background
- Alkane biosynthesis, as it is, is mainly limited by ADC’s activity. In an effort to diversify alkane production, we strived to improve ADC’s activity on shorter fatty aldehydes.
- A crystal structure derived model of ADC was put on a Pymol file and reveals a octadecanoic acid stuck in the protein. The octadecanoic acid was modified to a tetradecanal, and then the Pymol file was converted to a Fold-it file for human interaction. On Fold-it, we can model mutations of amino acids in the enzyme and Fold-it computes a relative value representing the enzyme’s stability. As we wanted to maintain the aldehyde decarbonylation, we avoided changing amino acids near the catalytic iron center that binds to the aldehyde group.
- The most promising mutation sites seem to be amino acids 21 to 25 and 67 to 71, sections of two α-helices adjacent to the tetradecanal's carbon end; by mutating those amino acids to ones with larger side-chains, tetradecanal might fit better in ADC.
Methods
After using Fold-It, a computational protein design program, to model several ADC mutants, the ADC mutants were made using Kunkel Mutagenesis. The sequences of these mutants were later verified to carry the correct mutations. After creating several ADC mutants, three different types of assays were developed and the mutants were later tested and analyzed using Gas chromatography–mass spectrometry (GC-MS) to see if our mutants can produce more C13 alkanes. Three different assays were set up: cell lysis assay, protein purification assay, and in vivo assay.
In Vitro Assays
In these assays, ADC was cloned into a vector with an inducible promoter and Kanamycin resistance.
- Cell Lysis Assay
One aldehyde decarbonylase (ADC) colony was picked and grown in the 37°C shaker until OD600 reached the value of 0.8. Then, the culture was induced with IPTG and cell pellet was collected by spinning down after three more hours of shaking. This cell pellet was resuspended in a solution that contains sodium phosphate buffer, protease inhibitor, DNAses and lysozyme. The resuspended cells were lysed with 10% triton then centrifuged. The supernatant was transferred to a glass vial and used to set up in vitro reactions. In vitro reactions were set up by adding certain volume of cells, feredoxin, feredoxin reductase, NADPH and aldehyde. However, this assay did not yield consistent alkane production every time even though we kept variables very minimal. So our team decided to move on and develop a different assay, protein purification assay.
- Protein Purification Assay
We decided to increase the concentration of ADC and decrease the number of contaminants in the assay. We grow up cells to express 10 times more ADC proteins and then we purify the proteins by lysing the cells and binding the target ADC proteins to the BioRad columns and then eluting them out, resulting in pure ADC proteins. Then, we setted up in vitro reactions by adding purified ADC mutant protein, standard buffer(Hepes at pH8, NaCl, glycerol, TCEP and water) and feredoxin, feredoxin reductase, NADPH, and aldehyde into an eppendorf. After certain time period has passed, it was quenched in ethyl acetate and the top organic layer that results from quencing was extracted to an insert in a GC-MS vial. This was later analyzed in the GC-MS.
In Vivo Assay
We took the base construct RED-PSB1C3 (Acyl-ACP Reductase in PSB1C3 high constitutive vector) and made the 1C3-const-RED-rbs-mutant decarb construct (1C3 construct with RED, ribosomal binding site, and the mutant decarbonylase genes made with Kunkel (site directed) mutagenesis. Then we transformed the competent cells (BL-21) to take up construct plasmids with our mutated ADC genes. We then grow the transformed competent cells in TB with chloramphenicol and then plate it on TB-chloramphenicol agar plates. Individual colonies were taken to inoculate water, and the colony waters were taken to run colony PCRs to confirm the uptake of the correct plasmid constructs that are with mutated ADC genes. Once verified, the corresponding colony waters of the successful colonies were grown in TB-chloramphenicol media overnight. The media are then miniprepped to obtain DNA of the original plasmid construct taken up by BL-21. The miniprepped plasmid construct is then transformed into another form of the competent cells (MG1655) and then plated on TB-chloramphenicol agar to have samples of different genetic variety of samples. Six isolated colonies are picked on each plate (to average out genetic differences) to be grown in special alkane production media M9 for 48 hours. After that, alkane is extracted with ethyl acetate and the samples are then analyzed using GC-MS.
Current Status & Results
We have made and tested 5 mutant Aldehyde Decarbonylases. However, due to time constraints and insufficient protein storage conditions, we lack usable data from the purified protein assays. It's possible that ADC loses effectiveness quickly even when stored. From our experience, if one's sole purpose is to test mutants, the best thing to do is use a purified protein assay with C14 aldehydes to test improved activity, then characterize the wild type ADC and the promising mutants with C16 aldehyde assays and C18 aldehyde assays. An in vivo assay may work if LuxCDE constructs are made, as LuxCDE reportedly produces a C14 aldehyde.
Parts Submitted
We have submitted our five Aldehyde Decarbonylase mutants.
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590037 Aldehyde Decarbonylase_N25M]
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590038 Aldehyde Decarbonylase_C70V]
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590039 Aldehyde Decarbonylase_C70K]
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590040 Aldehyde Decarbonylase_Y21W]
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590040 Aldehyde Decarbonylase_Y21W, C70I]
Below are the submitted parts that co-express Aldehyde Decarbonylase(normal or mutant) and Acyl-ACP Reductase, the two genes responsible for alkane production.
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590053 RrD_PSB1C3-High constitutive]
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590054 RrD_N25M-PSB1C3-High constitutive]
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590055 RrD_C70V-PSB1C3-High constitutive]
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590056 RrD_C70K-PSB1C3-High constitutive]
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590057 RrD_Y21W-PSB1C3-High constitutive]
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590058 RrD_Y21W, C70I-PSB1C3-High constitutive]
 "
"