Team:Arizona State/Parts
From 2011.igem.org
(Difference between revisions)
Ethan ward (Talk | contribs) |
|||
| (9 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{:Team: | + | {{:Team:Arizona State/Templates/main|title=Parts|content= |
| + | | ||
| + | <!-- need padding here or the icon in the content panel will overlap the groupparts widget. --> | ||
| - | == | + | == BBa_K578001 == |
| + | '''E. coli CRISPR I leader sequence''' | ||
| - | + | This is the AT-rich leader region of the CRISPR I array in Escherichia coli str. K-12 substr. MG1655. "Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS" (Ü. Pul et al, 2010) characterized a single promoter (Pcrispr1) in this sequence. Transcription is silenced through H-NS binding to the leader sequence. Induction requires anti-silencing, either by protein-independent processes, DNA-binding proteins or other modulatory mechanisms not yet identified. | |
| + | == BBa_K578002 == | ||
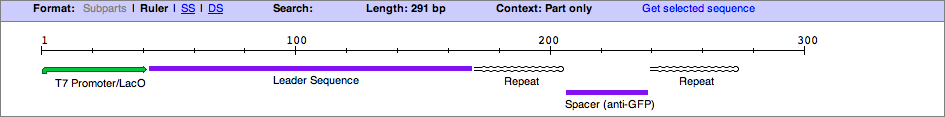
| + | '''T7LRSR (Listeria innocua CLIP11262)''' | ||
| + | [[image: BBa_K578002.png|800px]] | ||
| - | <groupparts> | + | This brick, T7LRSR, is the starting point for a CRISPR (Clustered Regularly Interspaced Short Palindromic Repeat) array based around Listeria Innocua CLIP11262. The sequence of the part is composed of a T7 promoter, Lac Operator, Leader Sequence (from Listeria Innocua CLIP11262), Palindromic Repeat, Spacer (Targeted to sequence with correct protospacer and 'NGG; Protospacer-adjacent motif), followed by a palindromic repeat. After the aforementioned sequences there is a pseudospacer with the standard BioBrick suffix (with SpeI and PstI Sites). The suffix allows for parts (eg.Repeat.Spacer Repeat, RSR, formated BioBricks) to be added onto the distal end of the CRISPR array. |
| + | == BBa_K578003 == | ||
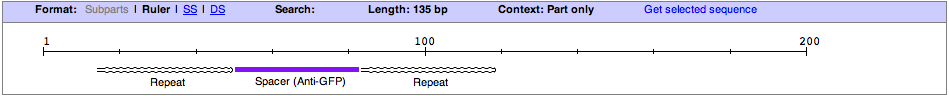
| + | '''RSR Array Segment (Listeria innocua CLIP11262)''' | ||
| + | |||
| + | [[image: BBa_K578003.png|800px]] | ||
| + | |||
| + | This brick, T7LRSR, is the starting point for a CRISPR (Clustered Regularly Interspaced Short Palindromic Repeat) array based around Listeria Innocua CLIP11262. The sequence of the part is composed of a T7 promoter, Lac Operator, Leader Sequence (from Listeria Innocua CLIP11262), Palindromic Repeat, Spacer (Targeted to sequence with correct protospacer and 'NGG; Protospacer-adjacent motif), followed by a palindromic repeat. After the aforementioned sequences there is a pseudospacer with the standard BioBrick suffix (with SpeI and PstI Sites). The suffix allows for parts (eg.Repeat.Spacer Repeat, RSR, formated BioBricks) to be added onto the distal end of the CRISPR array. | ||
| + | == BBa_K578004 == | ||
| + | '''Bacillus halodurans C-125 CRISPR leader sequence''' | ||
| + | |||
| + | This is a putative leader sequence for the CRISPR III locus in Bacillus halodurans C-125. | ||
| + | == BBa_K578005 == | ||
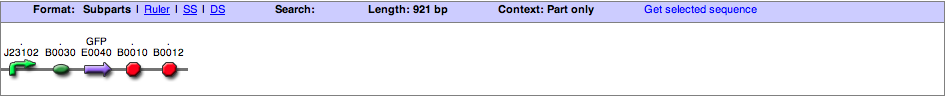
| + | '''J23102+E0840 (Constitutive GFP)''' | ||
| + | |||
| + | This is a combination of the GFP coding region (E0840), and the medium strength constitutive promoter, J23102. | ||
| + | [[image: ASU_BBa_K578005_2.png|800px]] | ||
| + | <!--<groupparts>iGEM11_Arizona_State</groupparts>--> | ||
| + | |||
| + | }} <!-- close content attribute for menubar --> | ||
Latest revision as of 03:12, 29 September 2011
 "
"