Team:Paris Bettencourt/T7 diffusion
From 2011.igem.org
| (34 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Paris_Bettencourt/tpl_test}} | {{:Team:Paris_Bettencourt/tpl_test}} | ||
| - | |||
| - | |||
<html> | <html> | ||
| - | The | + | <h1>The T7 RNA polymerase design</h1> |
| - | + | <table> | |
| + | <tr> | ||
| + | <td> | ||
| + | <p><em>Bacteriophage T7 RNA polymerase</em> is a DNA-dependant RNA polymerase from the T7 bacteriophage genome. The enzyme is composed of a single polypeptide chain of 880 amino acids. It catalyzes the processive polymerization of messenger RNA from nucleoside triphosphate precursors by using one strand of DNA as a template . This enzyme is known to have a stringent specificity for its promoter, that is orthogonal to the other promoters of the cell.</p> | ||
| + | <p>In our designs, we wanted a protein to pass through the tubes and trigger a signal in the receiver cell. T7 RNA polymerase is considered as a very good candidate for such systems. This explains why it is used as the biggest<a href="https://2011.igem.org/Team:Paris_Bettencourt/Designs"> of our proof of principle molecules.</a></p> | ||
| + | </td> | ||
| + | <td> | ||
| + | | ||
| + | </td> | ||
| + | <td> | ||
| + | <center> | ||
| + | <img src="http://upload.wikimedia.org/wikipedia/commons/e/e3/T7_RNA_polymerase.jpg" width=200px><br/> | ||
| + | <b><u>Fig1:</u></b> Cristallographic structure of RNA polymerase. <a ref="http://en.wikipedia.org/wiki/T7_RNA_polymerase">(source)</a></center> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | |||
| + | <h2>Making the T7 RNA polymerase diffuse through the tubes</h2> | ||
| + | |||
| + | <p>When we calculated the radius of the T7 RNA polymerase we noticed that it is smaller than the average size of the nanotubes we can measure from the Dubey and Ben-Yehuda paper. We made the hypothesis that such a big molecule has the chance to pass through the tubes and we started building the design to test it.</p> | ||
| + | |||
| + | <ul> | ||
| + | <li><em>In the emitter cell</em>, we have to over express the T7 polymerase. As we said in the <a href="https://2011.igem.org/Team:Paris_Bettencourt/Designs">general overview</a> the production of T7 polymerase is under the control of an IPTG inducible promoter design. The RFP, placed on the same mRNA is a reporter of the quantity of the produced T7 polymerase.</li> | ||
| + | |||
| + | <li><em>In the receiver cell</em>, a system sensitive to the T7 polymerase will be activated if a T7 polymerase reaches its promoter, present in a few plasmids of the receiver cell (low copy plasmids). The system is self-amplifying and the GFP is produced as a monitor of the signal.</li> | ||
| + | </ul> | ||
| + | <p>The principles of the design are summed up in the image below:</p> | ||
<br/> | <br/> | ||
| - | |||
| + | <center> | ||
| + | <img src="https://static.igem.org/mediawiki/2011/9/99/T7_diffusion.jpg" width=800px/> | ||
| + | </p><b><u>Fig2:</u></b> Schematic summary of the T7 diffusion device</p> | ||
| + | </center> | ||
| + | |||
| + | |||
| + | <h2>T7 polymerase as an auto-amplifier</h2> | ||
| + | |||
| + | <p>One of our first concerns was the potential leaking from the auto-amplifier. Biological systems tend to be noisy. The promoter is known to be very orthogonal to the metabolism of the host. However, we had to deal with several problems. We invite you to see the experiments page for data about this.</p> | ||
| + | |||
| + | <p>We could not work in a synthetic biology plasmid since it does not exist for <i>B.subtilis</i>. We had to create and use a home made, non biobricked multihost plasmid, not designed to be silent. We had to add several terminators before the pT7 promoters in order to prevent the leakage.</p> | ||
| + | |||
| + | <p>As an alternative solution, we also constructed a GFP with a T7 promoter. This system is supposed to have a lower response, but without auto-amplification, leaking should be reduced to a minimum.</p> | ||
| + | |||
| + | <h2>Model and experiments</h2> | ||
| + | |||
| + | <p>To know more about what we have done with this system, we invite you to visit the corresponding <em>modeling</em> and <em>experiment</em> pages:</p> | ||
| + | <ul> | ||
| + | <li><em><a href="https://2011.igem.org/Team:Paris_Bettencourt/Modeling/T7_diffusion">Modeling</a></em></li> | ||
| + | <li><em><a href="https://2011.igem.org/Team:Paris_Bettencourt/Experiments/T7_diffusion">Experiments</a></em></li> | ||
| + | </ul> | ||
| + | <br/> | ||
<br/> | <br/> | ||
| - | |||
<!-- PAGE FOOTER -- ITEMS FROM COLUMN ! HAVE BEEN MOVED HERE -- RDR --> | <!-- PAGE FOOTER -- ITEMS FROM COLUMN ! HAVE BEEN MOVED HERE -- RDR --> | ||
| Line 67: | Line 112: | ||
</div> | </div> | ||
</body> | </body> | ||
| + | <div id="scroll_right"><a href="https://2011.igem.org/Team:Paris_Bettencourt/Modeling/T7_diffusion"><img src="https://static.igem.org/mediawiki/2011/e/e0/Arrow-right-big.png" style="width:100%;"></a><a href="https://2011.igem.org/Team:Paris_Bettencourt/Modeling/T7_diffusion">Model for T7</a></div> | ||
| + | |||
| + | |||
</html> | </html> | ||
Latest revision as of 01:40, 22 September 2011

The T7 RNA polymerase design
|
Bacteriophage T7 RNA polymerase is a DNA-dependant RNA polymerase from the T7 bacteriophage genome. The enzyme is composed of a single polypeptide chain of 880 amino acids. It catalyzes the processive polymerization of messenger RNA from nucleoside triphosphate precursors by using one strand of DNA as a template . This enzyme is known to have a stringent specificity for its promoter, that is orthogonal to the other promoters of the cell. In our designs, we wanted a protein to pass through the tubes and trigger a signal in the receiver cell. T7 RNA polymerase is considered as a very good candidate for such systems. This explains why it is used as the biggest of our proof of principle molecules. |
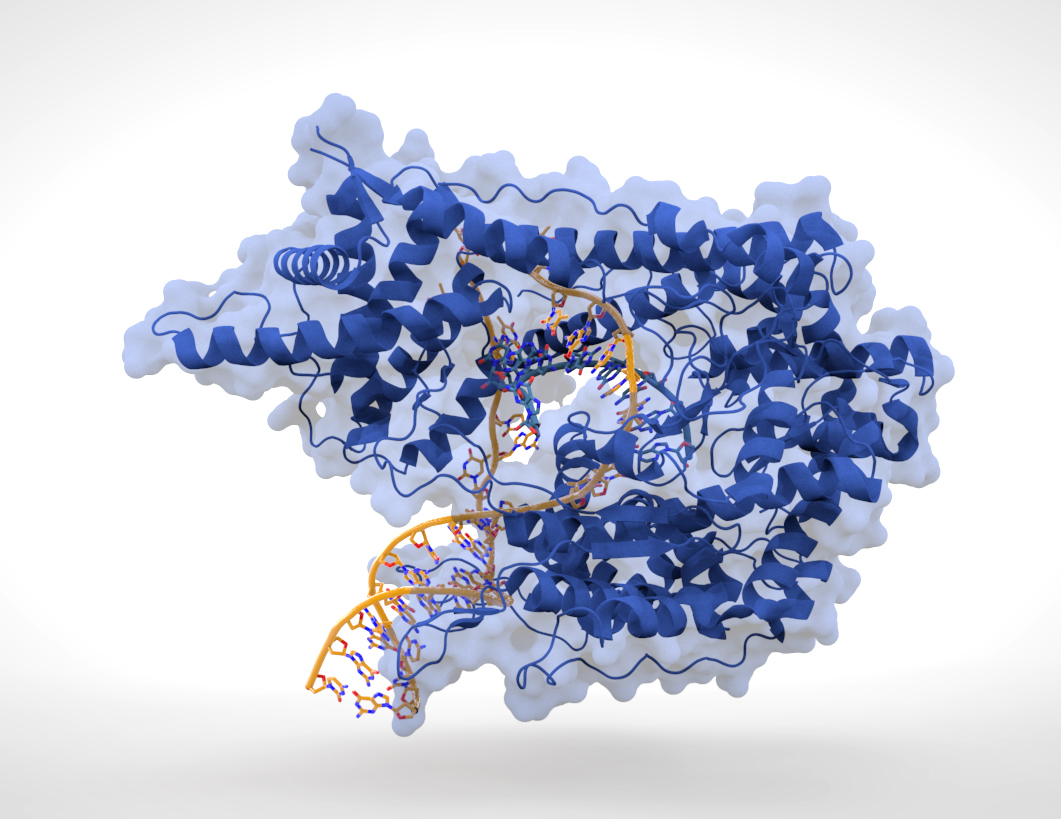 Fig1: Cristallographic structure of RNA polymerase. (source) |
Making the T7 RNA polymerase diffuse through the tubes
When we calculated the radius of the T7 RNA polymerase we noticed that it is smaller than the average size of the nanotubes we can measure from the Dubey and Ben-Yehuda paper. We made the hypothesis that such a big molecule has the chance to pass through the tubes and we started building the design to test it.
- In the emitter cell, we have to over express the T7 polymerase. As we said in the general overview the production of T7 polymerase is under the control of an IPTG inducible promoter design. The RFP, placed on the same mRNA is a reporter of the quantity of the produced T7 polymerase.
- In the receiver cell, a system sensitive to the T7 polymerase will be activated if a T7 polymerase reaches its promoter, present in a few plasmids of the receiver cell (low copy plasmids). The system is self-amplifying and the GFP is produced as a monitor of the signal.
The principles of the design are summed up in the image below:
 Fig2: Schematic summary of the T7 diffusion device
Fig2: Schematic summary of the T7 diffusion device
T7 polymerase as an auto-amplifier
One of our first concerns was the potential leaking from the auto-amplifier. Biological systems tend to be noisy. The promoter is known to be very orthogonal to the metabolism of the host. However, we had to deal with several problems. We invite you to see the experiments page for data about this.
We could not work in a synthetic biology plasmid since it does not exist for B.subtilis. We had to create and use a home made, non biobricked multihost plasmid, not designed to be silent. We had to add several terminators before the pT7 promoters in order to prevent the leakage.
As an alternative solution, we also constructed a GFP with a T7 promoter. This system is supposed to have a lower response, but without auto-amplification, leaking should be reduced to a minimum.
Model and experiments
To know more about what we have done with this system, we invite you to visit the corresponding modeling and experiment pages:
 "
"

