Team:EPF-Lausanne/Protocols/TetR
From 2011.igem.org
(Difference between revisions)
(→Protocol) |
(→PCR Cycle) |
||
| Line 20: | Line 20: | ||
== Protocol == | == Protocol == | ||
== PCR Cycle == | == PCR Cycle == | ||
| + | |||
<b>1.</b> 98°C for 30s | <b>1.</b> 98°C for 30s | ||
<b>2.</b> 98°C for 7.5 seconds | <b>2.</b> 98°C for 7.5 seconds | ||
<b>3.</b> 58°C for 20 sec (this depends on the primers, therefore look up the annealing temperature of the primers used) | <b>3.</b> 58°C for 20 sec (this depends on the primers, therefore look up the annealing temperature of the primers used) | ||
| - | <b>4.</b> 72°C for 15 sec; you have to calculate the time required. This polymerase has an effciency of 15-30s/kb. (calculate for the longest fragment) | + | <b>4.</b> 72°C for 15 sec; you have to calculate the time required. This polymerase has an effciency of 15-30s/kb. |
| + | (calculate for the longest fragment) | ||
<b>5.</b> 72°C for 5 min | <b>5.</b> 72°C for 5 min | ||
<b>6.</b> 4°C "forever" | <b>6.</b> 4°C "forever" | ||
| + | |||
*Save the file in the MAIN folder→ OK→ SAVE→RUN→Select the block you use→ RUN→ Change sample volume to 50µl, LidT should be 105°C and the box just below ("hotlid") should ALWAYS be checked | *Save the file in the MAIN folder→ OK→ SAVE→RUN→Select the block you use→ RUN→ Change sample volume to 50µl, LidT should be 105°C and the box just below ("hotlid") should ALWAYS be checked | ||
*Press STATUS to check the progress | *Press STATUS to check the progress | ||
Revision as of 12:24, 7 July 2011
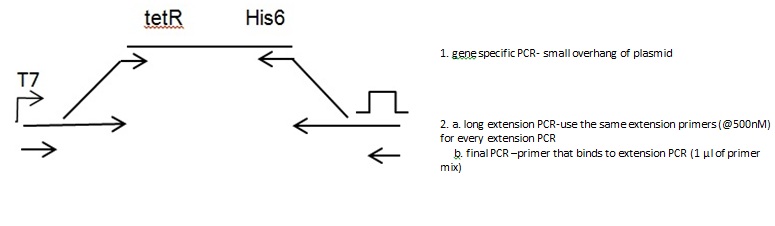
Linear template- TetR
Two step extension PCR
Contents |
Gene PCR
Materials
- 10µl 5x iProof HF buffer
- 0.5 µl of each primer
- 1 µl dNTP mix of 10mM
- 0.5 µl High Fidelity Polymerase
- 1 µl DNA template
- 0.5 µl Polymerase
- 36.5 µl H2O
Protocol
PCR Cycle
1. 98°C for 30s 2. 98°C for 7.5 seconds 3. 58°C for 20 sec (this depends on the primers, therefore look up the annealing temperature of the primers used) 4. 72°C for 15 sec; you have to calculate the time required. This polymerase has an effciency of 15-30s/kb. (calculate for the longest fragment) 5. 72°C for 5 min 6. 4°C "forever"
- Save the file in the MAIN folder→ OK→ SAVE→RUN→Select the block you use→ RUN→ Change sample volume to 50µl, LidT should be 105°C and the box just below ("hotlid") should ALWAYS be checked
- Press STATUS to check the progress
- Save the file in the MAIN folder→ OK→ SAVE→RUN→Select the block you use→ RUN→ Change sample volume to 50µl, LidT should be 105°C and the box just below ("hotlid") should ALWAYS be checked
- Press STATUS to check the progress
 "
"