Team:Paris Bettencourt/Experiments/ComS diffusion
From 2011.igem.org
Danyel.lee90 (Talk | contribs) |
|||
| (29 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
<html> | <html> | ||
<h1>ComS/ComK switch system</h1> | <h1>ComS/ComK switch system</h1> | ||
| - | |||
| - | <p>The ComS | + | <p>The ComS/ComK switch device is based on a bistable system known as the 'MeKS' module (interactions among the MecA complex, comK and comS). ComS inhibits the MecA protease and allows the ComK protein to self-amplify. But as ComK inhibits the production of ComS, the system comes back to the original state within a few hours. |
| - | < | + | <h2>Abstract</h2> |
| - | <p> | + | <div style="margin-left:50px; margin-right:50px; padding: 5px; border:2px solid black;"><b><p>Results for the ComK/ComS switch system: |
| + | <ul> | ||
| + | <li>We successfully BioBricked both the ComS gene (<a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K606037">BBa_K606037</a>) and the ComS gene codon optimized (<a href="http://partsregistry.org/Part:BBa_K606038">BBa_K606038</a>) constructs and sent them to the registry</li> | ||
| + | <li>We successfully knocked-out CodY gene in <i>B.subtilis</i></li> | ||
| + | <li>We characterized the ∆CodY <i>B.subtilis</i> strain</li> | ||
| + | <li>We saw that the system does not behave as our <a href="https://2011.igem.org/Team:Paris_Bettencourt/Modeling/ComS_diffusion">model</a> predicted, with a YFP-noise level lower than expected</li> | ||
| + | </ul></p></b></div> | ||
| + | <br/> | ||
| - | <h2> | + | <h2>Design overview</h2> |
| - | < | + | <center> |
| + | <img src="https://static.igem.org/mediawiki/2011/7/7f/ComS_summary.jpg" width=700px> | ||
| + | <p><u><b>Fig1:</b></u> Schematic of the ComS design</p> | ||
| + | </center> | ||
| + | <p>More information on the design <a href="https://2011.igem.org/Team:Paris_Bettencourt/ComS_diffusion">here</a>.</p> | ||
| - | <p> | + | <h2>Parts and biobrick system construction</h2> |
| + | <h3>ComS constructions</h3> | ||
| + | <p>The ComS gene was synthetized by GeneArt but also insulated by PCR colony, changing the start and the stop codon, and then cloned in pSB1C3. We then cloned in front of a pVeg-SpoVG promoter.</p> | ||
| - | < | + | <p>Here is the cloning plan we followed. Unfortunately, we didn't have the time to go until the B. Subtilis plasmid.</p> |
| + | |||
| + | <br /> | ||
| + | <br /> | ||
| + | <center><a href="https://static.igem.org/mediawiki/2011/4/40/1028_Cloning_plans_ComS_cloningPlan.png" ><img src="https://static.igem.org/mediawiki/2011/4/40/1028_Cloning_plans_ComS_cloningPlan.png" width=550></center></a> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | |||
| + | <h3>Creation of the ∆CodY strain with the reporter</h3> | ||
| + | |||
| + | <p>The genomic DNA from the CodY::spc strain from Linc Sonensheim laboratory was extracted and purified. The Elowitz's reporter strains were leaded to starvation, in order to make them competent, and then incubated for 40 min with the genomic DNA. During this time lapse, the strain is randomly making recombination between the extracellular DNA and its own genome, leading, after selection, to the KO strain.</p> | ||
| + | |||
| + | <p>Then, the B. Subtilis are selected on Spectinomycine plates, and only the KO clones were selected. This protocol works very well and we got a huge number of recombinants.</p> | ||
| - | <p> | + | <h2>Characterization of the ∆CodY strain</h2> |
| + | <p>We characterized ∆CodY B. subtilis strain.</p> | ||
| + | </html> | ||
| + | {| border="1" class="wikitable" style="text-align: center;" | ||
| + | |+∆CodY in B.subtilis at 37°C in exponential phase | ||
| + | |- | ||
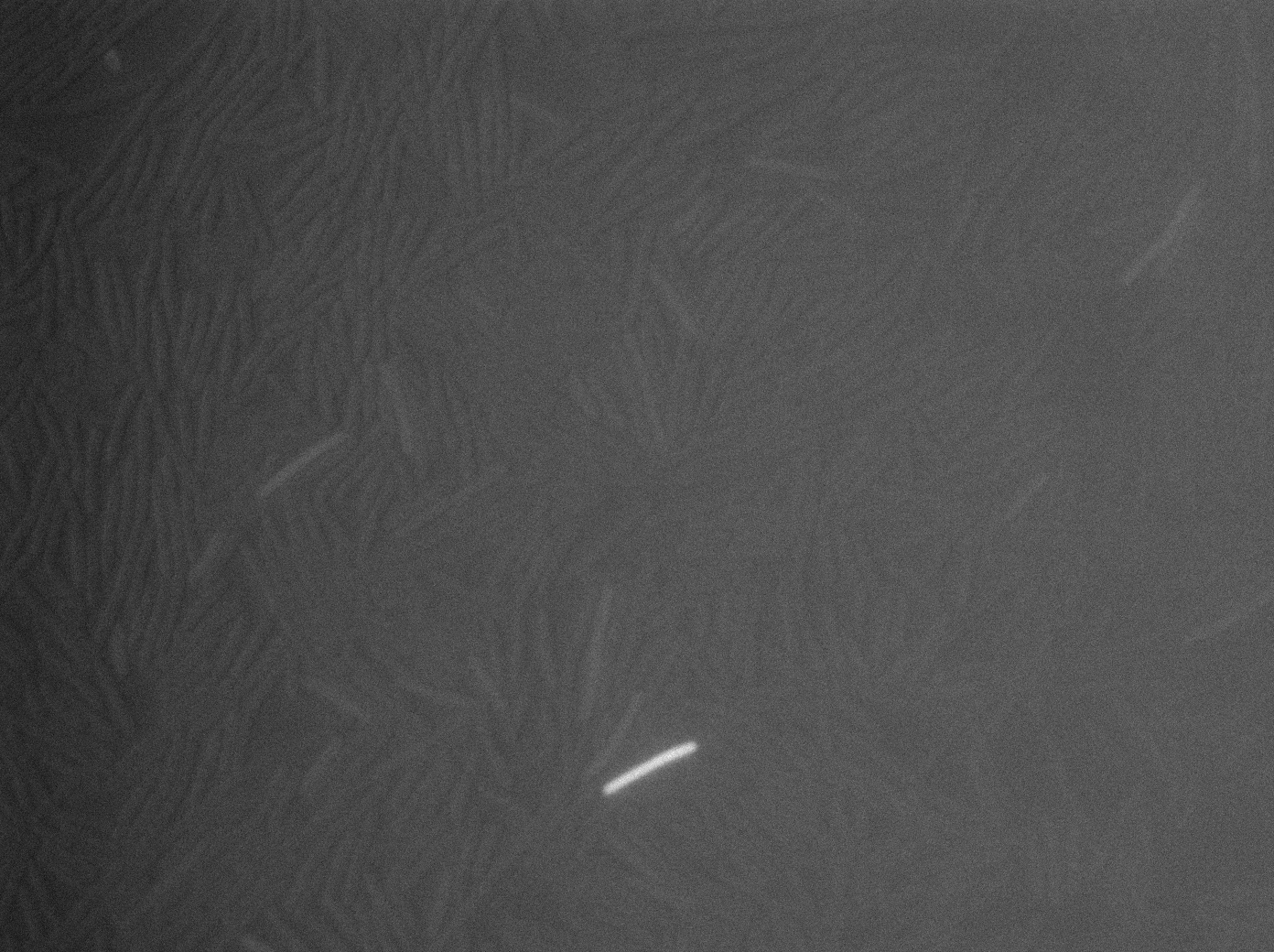
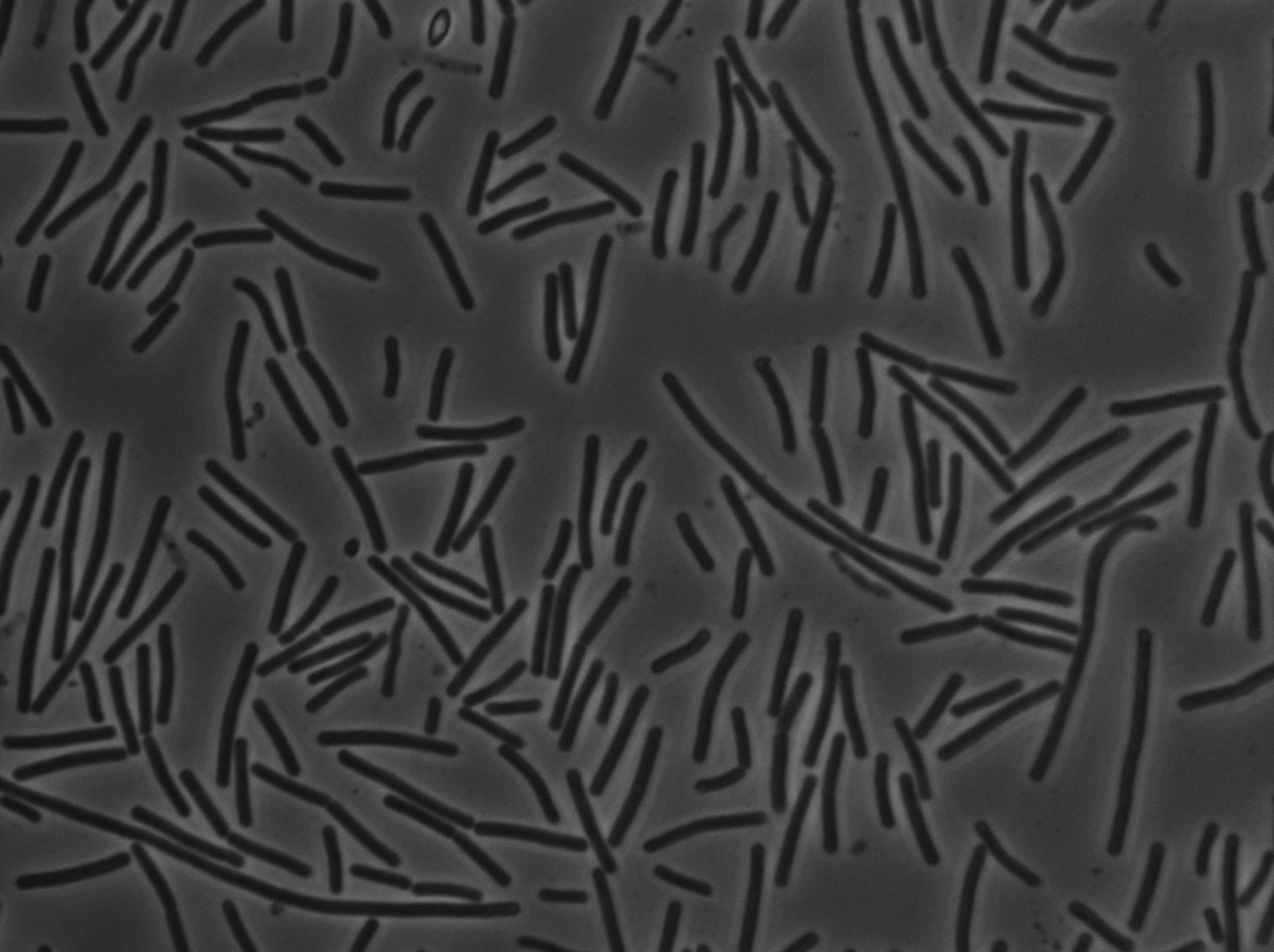
| + | |[[File:codY- trans s1t3 23102011.png|293px|thumb|center|B.subtilis ∆CodY at 37°C (trans image)]] | ||
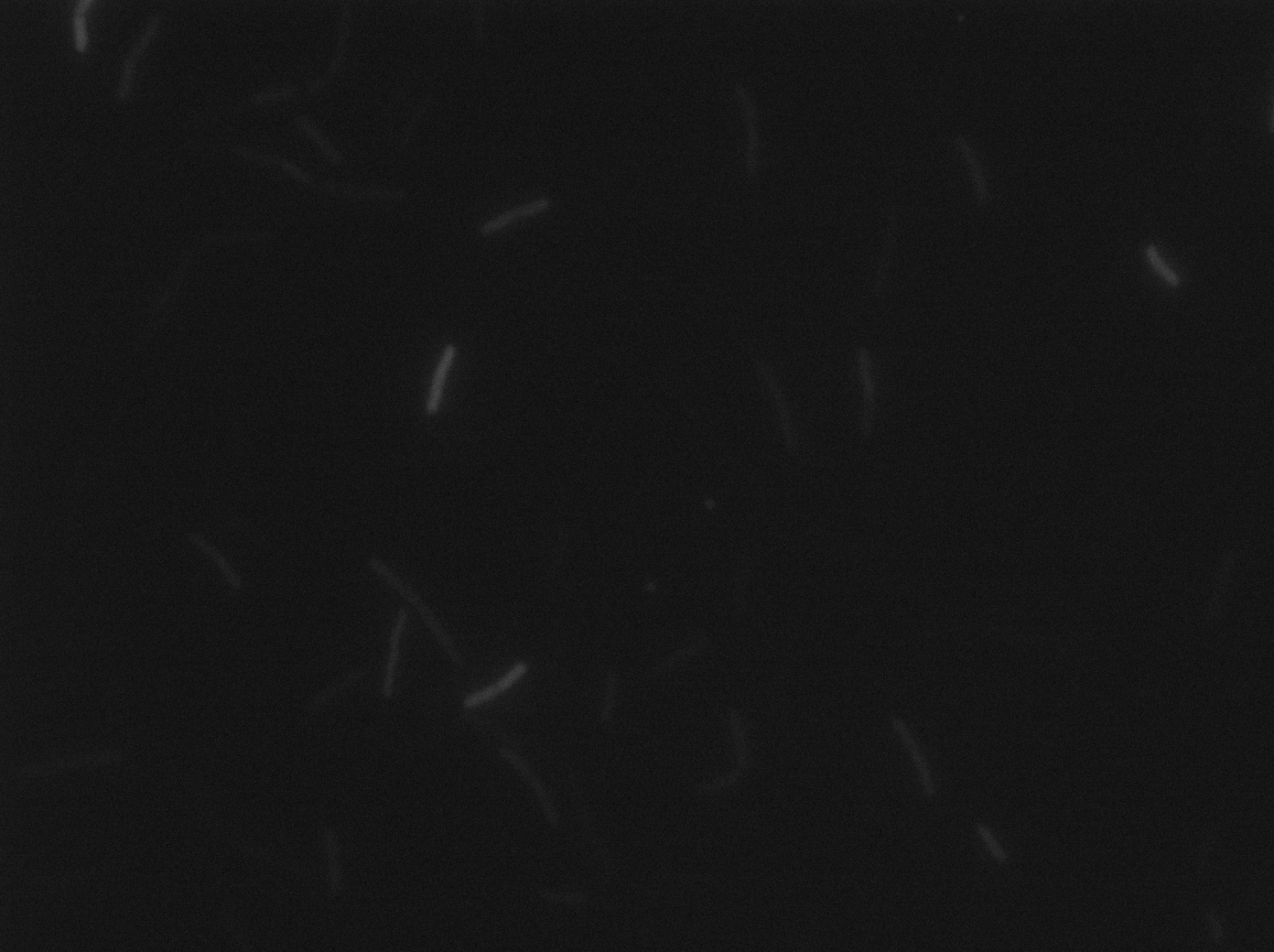
| + | |[[File:cody- yfp s1t3 23102011.png|293px|thumb|center|B.subtilis ∆CodY at 37°C (yfp image)]] | ||
| + | |[[File:cody- cfp s1t3 23102011.png|293px|thumb|center|B.subtilis ∆CodY at 37°C (cfp image)]] | ||
| + | |} | ||
| + | <html> | ||
| + | <br> | ||
| + | </html> | ||
| + | {| border="1" class="wikitable" style="text-align: center;" | ||
| + | |+Py79 control strain with the reporter php13-v10-6b at 37°C in exponential phase <html>(see the paper: <a href="https://2011.igem.org/Team:Paris_Bettencourt/ComS_diffusion#references">[2]</a>)</html> | ||
| + | |- | ||
| + | |[[File:py79ph10 trans s3t1.png|293px|thumb|center|B.subtilis PY79 at 37°C (trans image)]] | ||
| + | |[[File:cpy79ph10 yfp s3t1.png|293px|thumb|center|B.subtilis PY79 at 37°C (yfp image)]] | ||
| + | |[[File:py79ph10 cfp s3t1.png|293px|thumb|center|B.subtilis PY79 at 37°C (cfp image)]] | ||
| + | |} | ||
| + | <html> | ||
| + | </br> | ||
| + | <p>CodY is known to repress many genes of the steady state phase during the exponential phase. In order to avoid this problem, we have created a B.subtilis strain ∆CodY. This will allow the MeKS system to be active during the exponential growth phase, as well as thousands of stationnary phase genes. This mutation is not lethal.</p> | ||
| + | <p>During the exponential phase, in the ∆CodY strain, the system is supposed to be active, and can be stochastically switch on. Hence, we expect to find some cells in which the YFP and the CFP genes are expressed. There is a time lag between the activation of ComS and ComK that is longer than the time of our observations, so that there would be only one fluorescence visible at the time in a cell.</p> | ||
| + | <p>In the strain of Elowitz (PY79 B. subtilis with pHP13::PcomG-cfp-PcomS-yfp), CodY is expressed, which should block the system MeKS in exponential phase. Thus, we do not expect to see any fluorescence.</p> | ||
| + | <p>Let us first focus on YFP images. We see that in the population of the ∆CodY strain, there are few cells that express YFP. Surprisingly, we observe more YFP expression in CodY+ strain ( Elowitz' strain). Secondly, we clearly see that the ∆CodY strain does not express CFP apart from a bit of leakage. In the CodY+ strain, we can observe some fluorescent (CFP) bacterias as well. There are not enough example to derive a clear conclusion, but it seems to be that there are not significantly more CFP cells in the ∆CodY and in the CodY.</p> | ||
| + | <p>From this experiment, there is no clear evidences that the activity of the MeKS switch is recovered by the knock-out. There would be sevral possibilities that would explain these facts: the knock-out of the strain failed but we get the resistant strain to the expected antibiotic. Or we do not observe little fluorescence in the ∆CodY strain although we were expecting more activated cells in the ∆CodY strain. Though, we could wonder if the bistability of the system in exponential phase is indeed recovered.</p> | ||
| + | <p>This experiment relies on the stochastic activation of a really rare event. That is to say maybe the microscopy is not the better technique there. We can think about increasing the probability for the switch to turn on by expressing a high level of ComS within the cell, or by testing directly the competence, by transforming in these strains with the same amount of DNA and to count the colonies. We could also think about longer time experiments under the microscope, but we have to avoid the cells to enter in stationary phase and avoid multi layers.</p> | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
Latest revision as of 03:32, 29 October 2011

ComS/ComK switch system
The ComS/ComK switch device is based on a bistable system known as the 'MeKS' module (interactions among the MecA complex, comK and comS). ComS inhibits the MecA protease and allows the ComK protein to self-amplify. But as ComK inhibits the production of ComS, the system comes back to the original state within a few hours.
Abstract
Results for the ComK/ComS switch system:
- We successfully BioBricked both the ComS gene (BBa_K606037) and the ComS gene codon optimized (BBa_K606038) constructs and sent them to the registry
- We successfully knocked-out CodY gene in B.subtilis
- We characterized the ∆CodY B.subtilis strain
- We saw that the system does not behave as our model predicted, with a YFP-noise level lower than expected
Design overview

Fig1: Schematic of the ComS design
More information on the design here.
Parts and biobrick system construction
ComS constructions
The ComS gene was synthetized by GeneArt but also insulated by PCR colony, changing the start and the stop codon, and then cloned in pSB1C3. We then cloned in front of a pVeg-SpoVG promoter.
Here is the cloning plan we followed. Unfortunately, we didn't have the time to go until the B. Subtilis plasmid.

Creation of the ∆CodY strain with the reporter
The genomic DNA from the CodY::spc strain from Linc Sonensheim laboratory was extracted and purified. The Elowitz's reporter strains were leaded to starvation, in order to make them competent, and then incubated for 40 min with the genomic DNA. During this time lapse, the strain is randomly making recombination between the extracellular DNA and its own genome, leading, after selection, to the KO strain.
Then, the B. Subtilis are selected on Spectinomycine plates, and only the KO clones were selected. This protocol works very well and we got a huge number of recombinants.
Characterization of the ∆CodY strain
We characterized ∆CodY B. subtilis strain.
CodY is known to repress many genes of the steady state phase during the exponential phase. In order to avoid this problem, we have created a B.subtilis strain ∆CodY. This will allow the MeKS system to be active during the exponential growth phase, as well as thousands of stationnary phase genes. This mutation is not lethal.
During the exponential phase, in the ∆CodY strain, the system is supposed to be active, and can be stochastically switch on. Hence, we expect to find some cells in which the YFP and the CFP genes are expressed. There is a time lag between the activation of ComS and ComK that is longer than the time of our observations, so that there would be only one fluorescence visible at the time in a cell.
In the strain of Elowitz (PY79 B. subtilis with pHP13::PcomG-cfp-PcomS-yfp), CodY is expressed, which should block the system MeKS in exponential phase. Thus, we do not expect to see any fluorescence.
Let us first focus on YFP images. We see that in the population of the ∆CodY strain, there are few cells that express YFP. Surprisingly, we observe more YFP expression in CodY+ strain ( Elowitz' strain). Secondly, we clearly see that the ∆CodY strain does not express CFP apart from a bit of leakage. In the CodY+ strain, we can observe some fluorescent (CFP) bacterias as well. There are not enough example to derive a clear conclusion, but it seems to be that there are not significantly more CFP cells in the ∆CodY and in the CodY.
From this experiment, there is no clear evidences that the activity of the MeKS switch is recovered by the knock-out. There would be sevral possibilities that would explain these facts: the knock-out of the strain failed but we get the resistant strain to the expected antibiotic. Or we do not observe little fluorescence in the ∆CodY strain although we were expecting more activated cells in the ∆CodY strain. Though, we could wonder if the bistability of the system in exponential phase is indeed recovered.
This experiment relies on the stochastic activation of a really rare event. That is to say maybe the microscopy is not the better technique there. We can think about increasing the probability for the switch to turn on by expressing a high level of ComS within the cell, or by testing directly the competence, by transforming in these strains with the same amount of DNA and to count the colonies. We could also think about longer time experiments under the microscope, but we have to avoid the cells to enter in stationary phase and avoid multi layers.
 "
"