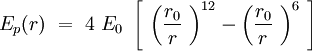Team:Paris Bettencourt/Modeling/Assisted diffusion/Membrane tension
From 2011.igem.org
| (27 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
<html> | <html> | ||
| - | <h1>Membrane tension calculation</h1> | + | <h1>2. Membrane tension calculation</h1> |
| + | |||
| + | <p> | ||
| + | Once the nanotube connexion between two bacteria is established ( click <a href='https://2011.igem.org/Team:Paris_Bettencourt/Modeling/Assisted_diffusion/Tube_formation'>here</a> to see the details on the nanotube formation ) what happens? Let's look on the membrane tension of two bacteria. | ||
| + | </p> | ||
<h2>Motivation</h2> | <h2>Motivation</h2> | ||
<p> | <p> | ||
| + | Let's evaluate the constraints that impact the membrane. As it is a Gram positive bacteria, the external sugar envelope imposes the shape of the bacteria. On the other hand, the osmotic pressure is pushing the membrane against the sugar wall. We assume that on the time scale of our model, the total number of phospholipids inside the membrane is fixed, but the density of phospholipids can fluctuate depending on the phospholipid production. So we can pretend that our system is evolving through a series of quasi-equilibrium states. | ||
| + | </p> | ||
| - | + | <html> | |
| + | <table> | ||
| + | <tr> | ||
| + | <td><center><img src="https://static.igem.org/mediawiki/2011/8/8a/MembraneCompression.png" height=200px></center></td> | ||
| + | <td><center><img src="https://static.igem.org/mediawiki/2011/d/d3/MembraneExtension.png" height=250px></center></td> | ||
| + | <tr> | ||
| + | <tr> | ||
| + | <td><p><center><u><b>Fig. 2:</b></u> If there is an excess of phospholipids, the membrane is being compressed<p></td> | ||
| + | <td><p><center><u><b>Fig. 3:</b></u> If there are not enough phospholipids, the membrane is being extended<p></td> | ||
| + | <tr> | ||
| + | </table> | ||
| + | </html> | ||
| + | <p> | ||
| + | The membrane tension is caused by interaction between phospholipids on the surface of bacteria. Closer the phospholipids are to each other, more the membrane is tense. | ||
</p> | </p> | ||
<p> | <p> | ||
| - | Can we describe the interaction between two phospholipids in the membrane? The question is complicated, but we will demonstrate that by describing properly the phospholipid, as it is reasonable to fit a Lennard-Jones potential energy to this interaction. | + | Can we describe the interaction between two phospholipids in the membrane? The question is complicated, but we will demonstrate that by describing properly the phospholipid, as it is reasonable to fit a Lennard-Jones potential energy to this interaction. Since the speed of conformational equilibration of phospholipid is way faster than the sliding of neighboring phospholipids we can consider phospholipids as cylinders (see the Fig. 4). To make things even simpler we decided to approximate these cylinders as points(see the Fig. 5). |
</p> | </p> | ||
| - | |||
<html> | <html> | ||
| Line 23: | Line 41: | ||
<td align="center" style="width:400px"><img src="https://static.igem.org/mediawiki/2011/e/e9/Image_phospholipide.png" height=200px></td> | <td align="center" style="width:400px"><img src="https://static.igem.org/mediawiki/2011/e/e9/Image_phospholipide.png" height=200px></td> | ||
<td align="center" style="width:400px"><img src="https://static.igem.org/mediawiki/2011/8/82/Mod%C3%A8le_Phospholipide.png" height=200px></td> | <td align="center" style="width:400px"><img src="https://static.igem.org/mediawiki/2011/8/82/Mod%C3%A8le_Phospholipide.png" height=200px></td> | ||
| + | <td align="center" style="width:400px"><img src="https://static.igem.org/mediawiki/2011/1/13/Point_ParisBettencourt.png" height=200px></td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td align="center"><center>Chemical structure of | + | <td align="center"><center><u><b>Fig. 3:</b></u> Chemical structure of a phospholipid</center></td> |
| - | <td align="center"><center> | + | <td align="center"><center><u><b>Fig. 4:</b></u> Cylindrical approximation of Israelachvili for a phospholipid</center></td> |
| + | <td align="center"><center><u><b>Fig. 5:</b></u> Our approximation</center></td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| Line 33: | Line 53: | ||
</html> | </html> | ||
| + | <center><h3> | ||
| + | As we can see the modelers go always straight to the point :) | ||
| + | </h3></center> | ||
| + | |||
| + | <p> | ||
Speaking in terms of energy, once we have these cones, it is reasonable to fit a Lennard-Jones potential to this interaction. If the cones are interpenetrating, a steric repulsion keeps the molecules apart, and the hydrophobic interaction that joins one lipid with the other acts as an attractive force keeping the coherence of the membrane. We will discuss later about the value of the two coefficients. | Speaking in terms of energy, once we have these cones, it is reasonable to fit a Lennard-Jones potential to this interaction. If the cones are interpenetrating, a steric repulsion keeps the molecules apart, and the hydrophobic interaction that joins one lipid with the other acts as an attractive force keeping the coherence of the membrane. We will discuss later about the value of the two coefficients. | ||
| + | </p> | ||
[[File:LJpot.jpg|center|Lennard-Jones potential]] | [[File:LJpot.jpg|center|Lennard-Jones potential]] | ||
| Line 43: | Line 69: | ||
<ul> | <ul> | ||
<li>We deal with a system under fixed temperature, so we can reason in canonical ensemble terminology</li> | <li>We deal with a system under fixed temperature, so we can reason in canonical ensemble terminology</li> | ||
| - | <li>Every bacterium in this model has a perfectly spherical form, so each phospholipid on it has 4 degrees of freedom</li> | + | <li>Every bacterium in this model has a perfectly spherical form, so each phospholipid on it has 4 degrees of freedom (2 coordinates, 2 momentums per phospholipid)</li> |
| - | <li> | + | <li>We will consider phosphlipids equally distributed on the membrane and only the initial density of this distribution can change</li> |
</ul></p></b></div> | </ul></p></b></div> | ||
| Line 71: | Line 97: | ||
<p> | <p> | ||
<center> | <center> | ||
| - | <img src='https://static.igem.org/mediawiki/2011/2/27/Partition_function.png' style="height: | + | <img src='https://static.igem.org/mediawiki/2011/2/27/Partition_function.png' style="height:35px"> |
</center> | </center> | ||
</p> | </p> | ||
| Line 121: | Line 147: | ||
<p> | <p> | ||
<center> | <center> | ||
| - | <img src='https://static.igem.org/mediawiki/2011/8/87/Tension_2.png' style="height: | + | <img src='https://static.igem.org/mediawiki/2011/8/87/Tension_2.png' style="height:55px"> |
</center> | </center> | ||
</p> | </p> | ||
| + | <p> | ||
| + | The next step after the membrane tension calculation is the <a href='https://2011.igem.org/Team:Paris_Bettencourt/Modeling/Assisted_diffusion/From_membrane_tension_to_liquid_flux'>3. Pressure difference and Result section.</a> | ||
| + | </p> | ||
| + | <html> | ||
| + | <p> | ||
| + | Click here to come back to <a href='https://2011.igem.org/Team:Paris_Bettencourt/Modeling/Assisted_diffusion'>Assisted diffusion section.</a></p> | ||
| + | </html> | ||
| + | |||
| + | |||
| + | <html> | ||
<!-- PAGE FOOTER -- ITEMS FROM COLUMN ! HAVE BEEN MOVED HERE -- RDR --> | <!-- PAGE FOOTER -- ITEMS FROM COLUMN ! HAVE BEEN MOVED HERE -- RDR --> | ||
| Line 176: | Line 212: | ||
</div> | </div> | ||
| + | |||
| + | </html> | ||
| + | <html> | ||
| + | <div id="scroll_left"><a href="https://2011.igem.org/Team:Paris_Bettencourt/Modeling/Assisted_diffusion"><img src="https://static.igem.org/mediawiki/2011/0/0a/Arrow-left-big.png" style="width:100%;"></a><a href="https://2011.igem.org/Team:Paris_Bettencourt/Modeling/Assisted_diffusion">To come back to Assisted diffusion section</a></div> | ||
| + | <div id="scroll_right"><a href="https://2011.igem.org/Team:Paris_Bettencourt/Modeling/Assisted_diffusion/From_membrane_tension_to_liquid_flux"><img src="https://static.igem.org/mediawiki/2011/e/e0/Arrow-right-big.png" style="width:100%;"></a><a href="https://2011.igem.org/Team:Paris_Bettencourt/Modeling/Assisted_diffusion/From_membrane_tension_to_liquid_flux">To go to Pressure difference and Result section</a></div> | ||
</html> | </html> | ||
Latest revision as of 03:06, 29 October 2011

2. Membrane tension calculation
Once the nanotube connexion between two bacteria is established ( click here to see the details on the nanotube formation ) what happens? Let's look on the membrane tension of two bacteria.
Motivation
Let's evaluate the constraints that impact the membrane. As it is a Gram positive bacteria, the external sugar envelope imposes the shape of the bacteria. On the other hand, the osmotic pressure is pushing the membrane against the sugar wall. We assume that on the time scale of our model, the total number of phospholipids inside the membrane is fixed, but the density of phospholipids can fluctuate depending on the phospholipid production. So we can pretend that our system is evolving through a series of quasi-equilibrium states.
 |
 |
The membrane tension is caused by interaction between phospholipids on the surface of bacteria. Closer the phospholipids are to each other, more the membrane is tense.
Can we describe the interaction between two phospholipids in the membrane? The question is complicated, but we will demonstrate that by describing properly the phospholipid, as it is reasonable to fit a Lennard-Jones potential energy to this interaction. Since the speed of conformational equilibration of phospholipid is way faster than the sliding of neighboring phospholipids we can consider phospholipids as cylinders (see the Fig. 4). To make things even simpler we decided to approximate these cylinders as points(see the Fig. 5).
 |
 |
 |
As we can see the modelers go always straight to the point :)
Speaking in terms of energy, once we have these cones, it is reasonable to fit a Lennard-Jones potential to this interaction. If the cones are interpenetrating, a steric repulsion keeps the molecules apart, and the hydrophobic interaction that joins one lipid with the other acts as an attractive force keeping the coherence of the membrane. We will discuss later about the value of the two coefficients.
The membrane tension calculation
Some hypotheses used :
- We deal with a system under fixed temperature, so we can reason in canonical ensemble terminology
- Every bacterium in this model has a perfectly spherical form, so each phospholipid on it has 4 degrees of freedom (2 coordinates, 2 momentums per phospholipid)
- We will consider phosphlipids equally distributed on the membrane and only the initial density of this distribution can change



Using statistical physics approach, we can say that the phase space has 4N dimensions: two dimensions of impulsion and two dimensions of coordinate for each particle (N is the number of particles in the system).
where the Hamiltonian is :






The next step after the membrane tension calculation is the 3. Pressure difference and Result section.
Click here to come back to Assisted diffusion section.
 "
"