Team:Paris Bettencourt/Experiments/Methodologies/Integration
From 2011.igem.org
LauradaSilva (Talk | contribs) |
LauradaSilva (Talk | contribs) |
||
| (50 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Paris_Bettencourt/tpl_test}} | {{:Team:Paris_Bettencourt/tpl_test}} | ||
| - | <h1> | + | <h1>Integrative plasmids in <i>B. subtilis</i></h1> |
| - | Since | + | <h3>pDCPKO</h3> |
| + | |||
| + | Since cloning into B. subtilis with a replicative plasmid is not an easy task, we decided to construct an integrative plasmid which could give us a better efficiency. Moreover, we would observe less varialibity in gene expression than with a replicative plasmid. To achieve this goal, we designed and constructed a new biobricked integrative plasmid (pDCPKO) from pBGCS6 that we found in the <html><a href="http://www.bgsc.org/">Bacillus Genetic Stock Center</a></html>. | ||
{| border="1" class="wikitable" style="text-align: center;" | {| border="1" class="wikitable" style="text-align: center;" | ||
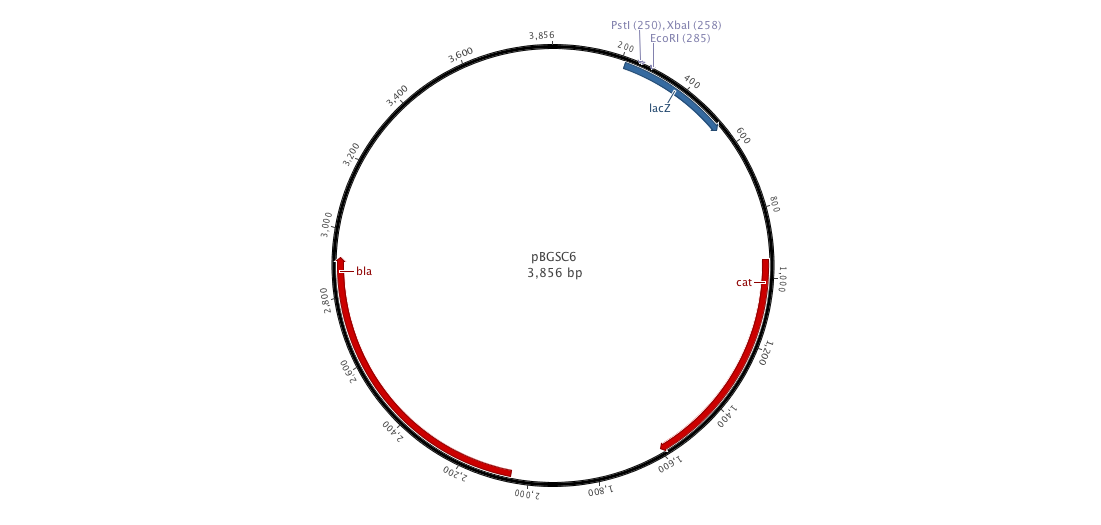
| - | |+ | + | |+ pBGCS6 (B. subtilis vector) |
|- | |- | ||
|[[File:pBGSC6.png|900px|center|]] | |[[File:pBGSC6.png|900px|center|]] | ||
| Line 13: | Line 15: | ||
|} | |} | ||
| - | <p>Into this | + | <p>Into this plasmid, we cloned both 3' and 5' AmyE sequences created by the 2008 Imperial College of London iGEM team (<partinfo>K143001</partinfo> and <partinfo>K143002</partinfo>). In the design of our new pDCPKO plasmid, we placed our AmyE sequences in order to integrate the whole plasmid into B.subtilis. pDCPKO encodes a chloramphenicol acetyl transferase (cat) selectable in either E.coli or B.subtilis (chloramphenicol 5μg/ml), β-lactamase (bla) selectable in E.coli only (ampicillin 100μg/ml).</p> |
{| border="1" class="wikitable" style="text-align: center;" | {| border="1" class="wikitable" style="text-align: center;" | ||
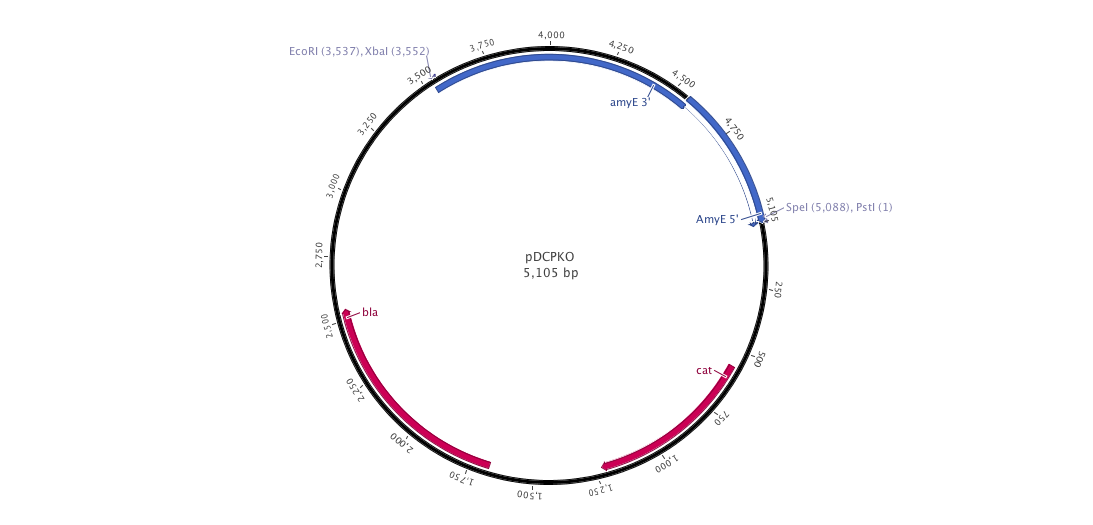
| - | |+ | + | |+ pDCPKO (B.subtilis integrative plasmid) |
|- | |- | ||
| - | |[[File:pDCPKO | + | |[[File:pDCPKO.png|900px|center|]] |
|- | |- | ||
|} | |} | ||
| - | <p> | + | <p>Here you can see that we managed to clone some of our parts into pDCPKO and transformed them into E.coli. |
| + | </p> | ||
| + | |||
{| border="1" class="wikitable" style="text-align: center;" | {| border="1" class="wikitable" style="text-align: center;" | ||
| - | |+ | + | |+pDCPKO in E.coli |
|- | |- | ||
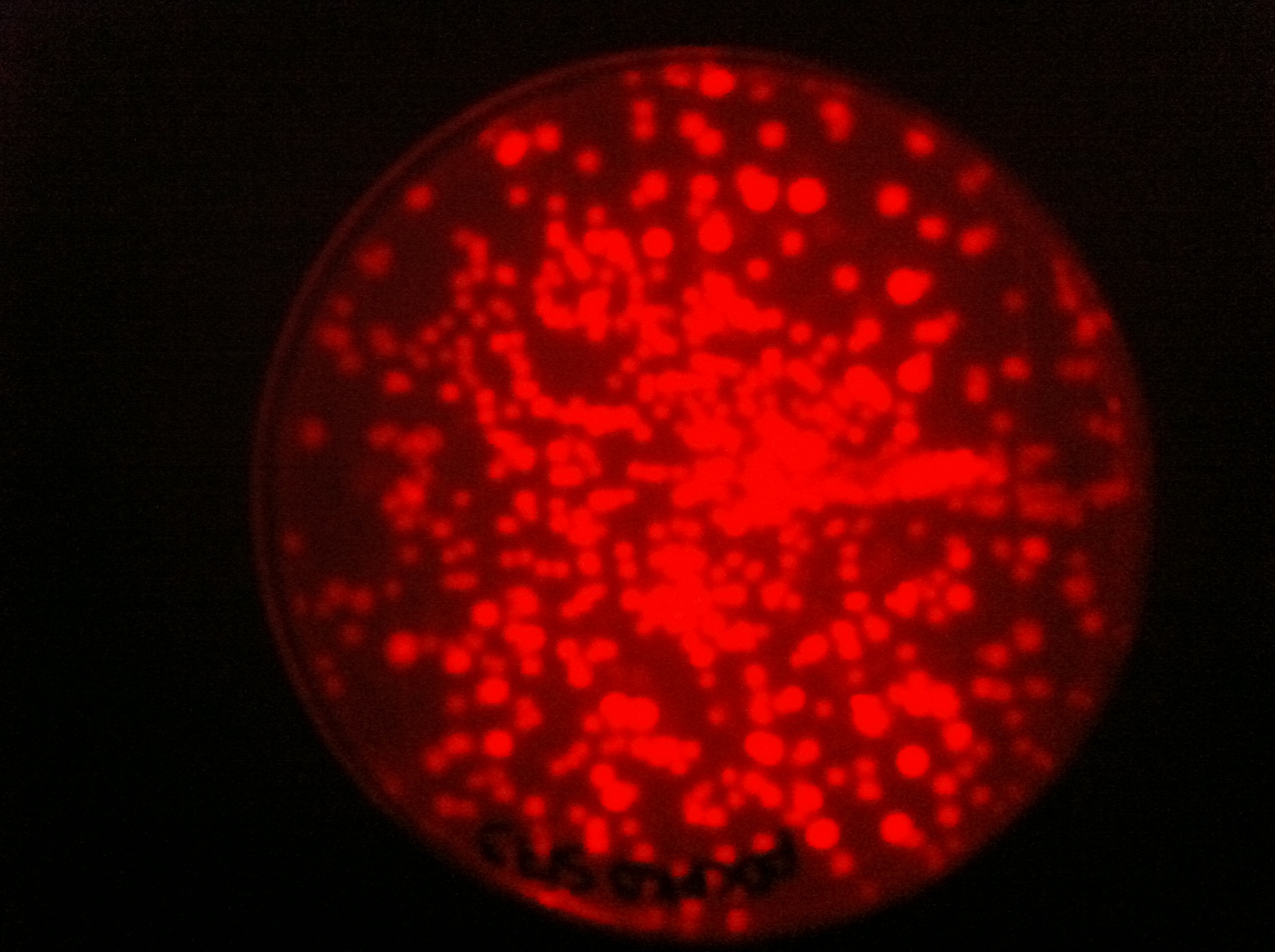
| - | |[[File:pDG364.png| | + | |[[File:pDCPKOs75ec1.jpg|450px|thumb|center|pDCPKO with RFP cassette in E.coli]] |
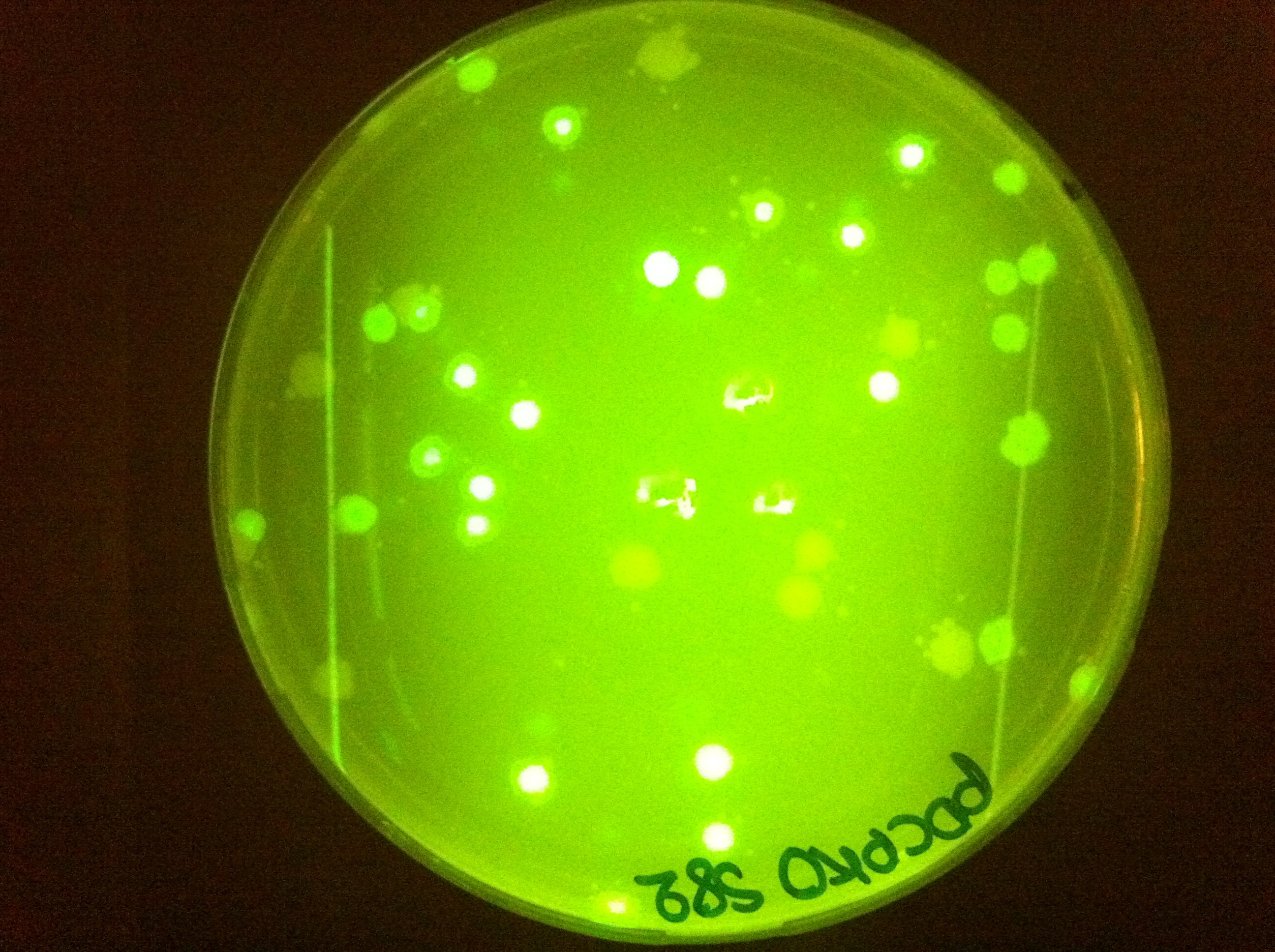
| + | |[[File:PDCPKOs82ec1.jpg|450px|thumb|center|pDCPKO with GFP cassette in E.coli]] | ||
| + | |} | ||
| + | |||
| + | <p>Until now, we are still awaiting results for the integration of pDCPKO plasmid into B.subtilis.</p> | ||
| + | |||
| + | <h3>pDG364</h3> | ||
| + | |||
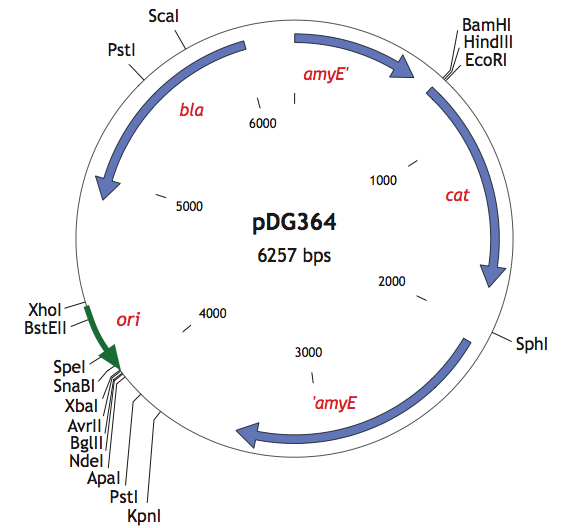
| + | <p>We also used the pDG364 integrative plasmid from Harald Putzer at <html><a href="http://www.ibpc.fr/UPR9073/AccueilUPR9073.htm">IBPC</a></html> which encodes a chloramphenicol acetyl transferase (cat) selectable in either E.coli or B.subtilis (chloramphenicol 5μg/ml), β-lactamase (bla) selectable in E.coli only (ampicillin 100μg/ml). | ||
| + | <center> | ||
| + | {| border="1" class="wikitable" style="text-align: center;" | ||
| + | |+ pDG364 (B.subtilis integrative vector) | ||
| + | |- | ||
| + | |[[File:pDG364.png|400px|center|]] | ||
|- | |- | ||
|} | |} | ||
| + | </center> | ||
| + | In order to integrate into this plasmid, we added BamHI restriction site to our biobricks using PCR. Thus, we were able to clone our parts into the pDG364 plasmid by cutting with BamHI and EcoRI.</p> | ||
| - | <p> | + | <p>Here you can see that we managed to clone some of our parts into pDG364 and transformed them into B.subtilis.</p> |
| - | + | {| border="1" class="wikitable" style="text-align: center;" | |
| - | + | |+pDG364 in E.coli and B.subtilis | |
| + | |- | ||
| + | |[[File:pDG364s75ec1.jpg|450px|thumb|center|pDG364 with RFP cassette in E.coli]] | ||
| + | |[[File:PDG364S75bs1.jpg|450px|thumb|center|pDG364 with RFP cassette in B.subtilis]] | ||
| + | |} | ||
| + | <html> | ||
| - | <h1>Starvation</h1> | + | <p>To transform our integrative plasmids (pDCPKO and pDG364) into B.subtilis, we chose two different methods: <b>starvation</b> and <b>electroporation</b>.</p> |
| + | |||
| + | <br> | ||
| + | |||
| + | |||
| + | <h1>Starvation protocol for integration into <i>B.subtilis</i></h1> | ||
| + | |||
| + | <p>The first one is the starvation method which is based on the natural competence in B.subtilis.</p> | ||
<p style="display:block;"><u>MDCH medium</u> | <p style="display:block;"><u>MDCH medium</u> | ||
</p> | </p> | ||
| Line 83: | Line 114: | ||
</li></ul> | </li></ul> | ||
| + | <br> | ||
| + | <h1>Electroporation protocol for integration into <i>B.subtilis</i></h1> | ||
| - | < | + | <p>The second method used to integrate into B.subtilis is based on electroporation.</p> |
<ul><li>Reagents and Equipment needed: | <ul><li>Reagents and Equipment needed: | ||
Mannitol, Sorbitol, Trehalose, LB, glycerol (99,5%) | Mannitol, Sorbitol, Trehalose, LB, glycerol (99,5%) | ||
| Line 92: | Line 125: | ||
Micropipettes: P2, P200, P1000 | Micropipettes: P2, P200, P1000 | ||
Pipettes: 25 mL, 10 mL, 5 mL</ul> | Pipettes: 25 mL, 10 mL, 5 mL</ul> | ||
| - | + | <br> | |
| - | + | <i>Day 1: preparation</i> | |
| - | + | <br> | |
<ul><li>Growth medium: LB + 0.5 mol.L-1 sorbitol → expected final volume: 52 mL for 1 cell culture (including 1 mL for taring the absorbance machine) | <ul><li>Growth medium: LB + 0.5 mol.L-1 sorbitol → expected final volume: 52 mL for 1 cell culture (including 1 mL for taring the absorbance machine) | ||
<p>_ msorbitol ≈ 4,736 g</p> </ul> | <p>_ msorbitol ≈ 4,736 g</p> </ul> | ||
| Line 110: | Line 143: | ||
<p>Inoculate a falcon containing 10 ml of LBA with your bacillus strain and let it grow overnight (37°C with shaking).</p> | <p>Inoculate a falcon containing 10 ml of LBA with your bacillus strain and let it grow overnight (37°C with shaking).</p> | ||
| - | + | <i>Day 2: electroporation</i> | |
| - | + | <br> | |
<p>1) Monitor the OD600 of your overnight culture</p> | <p>1) Monitor the OD600 of your overnight culture</p> | ||
<p>2) In a 500 ml erlenmeyer: dilute your culture into 50mL of Growth Medium so that the OD 600 is 0.01</p> | <p>2) In a 500 ml erlenmeyer: dilute your culture into 50mL of Growth Medium so that the OD 600 is 0.01</p> | ||
Latest revision as of 02:29, 29 October 2011

Integrative plasmids in B. subtilis
pDCPKO
Since cloning into B. subtilis with a replicative plasmid is not an easy task, we decided to construct an integrative plasmid which could give us a better efficiency. Moreover, we would observe less varialibity in gene expression than with a replicative plasmid. To achieve this goal, we designed and constructed a new biobricked integrative plasmid (pDCPKO) from pBGCS6 that we found in the Bacillus Genetic Stock Center.
Into this plasmid, we cloned both 3' and 5' AmyE sequences created by the 2008 Imperial College of London iGEM team (<partinfo>K143001</partinfo> and <partinfo>K143002</partinfo>). In the design of our new pDCPKO plasmid, we placed our AmyE sequences in order to integrate the whole plasmid into B.subtilis. pDCPKO encodes a chloramphenicol acetyl transferase (cat) selectable in either E.coli or B.subtilis (chloramphenicol 5μg/ml), β-lactamase (bla) selectable in E.coli only (ampicillin 100μg/ml).
Here you can see that we managed to clone some of our parts into pDCPKO and transformed them into E.coli.
Until now, we are still awaiting results for the integration of pDCPKO plasmid into B.subtilis.
pDG364
We also used the pDG364 integrative plasmid from Harald Putzer at IBPC which encodes a chloramphenicol acetyl transferase (cat) selectable in either E.coli or B.subtilis (chloramphenicol 5μg/ml), β-lactamase (bla) selectable in E.coli only (ampicillin 100μg/ml).
Here you can see that we managed to clone some of our parts into pDG364 and transformed them into B.subtilis.
To transform our integrative plasmids (pDCPKO and pDG364) into B.subtilis, we chose two different methods: starvation and electroporation.
Starvation protocol for integration into B.subtilis
The first one is the starvation method which is based on the natural competence in B.subtilis.
MDCH medium
- Phosphate-citrate buffer (10 x PC) 10 x concentrated stock solution contains per liter : K2H PO4 (anhydrous) 107 g; KH2 PO4 (anhydrous) 60 g; Trisodium citrate (5 H2O) 10 g.
- Dilute stock solution, check pH of 1 x PC buffer and adjust (if necessary) to pH=7. (1 x PC corresponds to Spizizen's salts without ammonium sulfate)
- '''10 ml''' MD medium : 1 x PC buffer 9.2 ml; Glucose (50 %, w/v) 0.4 ml; L-tryptophan (5 mg/ml) 0.1 ml; Ferric ammonium citrate (2.2 mg/ml) 0.05 ml; Potassium aspartate or potassium glutamate (100 mg/ml) 0.25 ml; 1 M MgSO4 0.03 ml.
- Add 0,2mL of casein hydrolysate in the MD medium
- A 10 ml preculture is grown overnight at room temperature in LM broth (liquid LB medium supplemented with auxotrophic requirements and 3 mM MgSO4). MDCH medium is prepared by adding 0.2 ml 5 % casein hydrolysate to 10 ml MD. Use the preculture to inoculate MDCH medium at an OD600 of about 0.05. Grow this culture at 37° C with shaking to To (transition from exponential to stationary phase). Sometimes, To do not appears and we can see a decrease of the OD: do not use the culture. Advice: launch 2 or 3 culture for each strain.
- Add 1 volume of fresh MD medium without casein hydrolysate to 1 volume of culture. Continue shaking at 37° C for 1 hour.
- Make aliquots of 500μL
- Add DNA (up to 1μg) and continue shaking in the presence of DNA for 20 minutes. Eventually, allow for expression of antibiotic resistance for 20mn (as described inJ. Bacteriol. (1988), 170 : 5093-5101) and plate.
- Cm: 5μg/mL; Spec: 50 to 100μg/mL; Kan: 5μg/mL, Erm: 1μg/mL '''with''' 25μg/mL of Lincomycin
Electroporation protocol for integration into B.subtilis
The second method used to integrate into B.subtilis is based on electroporation.
- Reagents and Equipment needed: Mannitol, Sorbitol, Trehalose, LB, glycerol (99,5%) DNA (50 ng/μL), B. subtilis strain for transformation Cuvette (2mm), Gene Pulser (Bio-rad) set on 200 ohms and 25 μF (≈ 5 ms pulses) and 2 to 2.5 kV Centrifuge set at 3000g and 10 minutes Micropipettes: P2, P200, P1000 Pipettes: 25 mL, 10 mL, 5 mL
Day 1: preparation
- Growth medium: LB + 0.5 mol.L-1 sorbitol → expected final volume: 52 mL for 1 cell culture (including 1 mL for taring the absorbance machine)
_ msorbitol ≈ 4,736 g
- Electro-poration medium: de-ionized water + 0.5 mol.L-1 sorbitol + 0.5 mol.L-1 mannitol + 0.5 mol.L-1 trehalose + 10% glycerol (v/v) → expected final volume: ≈ 40 mL for 1 round of poration
_ msorbitol = mmannitol ≈ 3,643 g
_ mtrehalose ≈ 7,566 g
_ Vglycerol (99,5 %) ≈ 4 mL
- Recovery medium: LB + 0.5 mol.L-1 sorbitol + 0.38 mol.L-1 mannitol → expected final volume: ≈ 1 ml per poly... tube (Approximately 10 tubes ≈ 11 ml total)
_ msorbitol ≈ 1,002 g
_ mmannitol ≈ 0,761 g
Sterilise the solution: Filtration or autoclave.
Inoculate a falcon containing 10 ml of LBA with your bacillus strain and let it grow overnight (37°C with shaking).
Day 2: electroporation1) Monitor the OD600 of your overnight culture
2) In a 500 ml erlenmeyer: dilute your culture into 50mL of Growth Medium so that the OD 600 is 0.01
3) Let the culture grow (37°C with shaking) until OD600 is between 0.85 and 1
4) Cool the cells on ice for 5 minutes.
5) NOTE: KEEP ALL YOUR MATERIAL ON ICE AND ALWAYS MANIPULATE ON ICE FROM NOW ON, KEEP AS STERILE AS POSSIBLE.
6) Distribute evenly the culture into two falcons and centrifuge at 3000g for 10 minutes.
7) Get rid of supernatant, tap the falcon upside down on a piece of paper and detach the pellet.
8) Add 20 mL of ice-cold electro-poration medium to one falcon, suspend the cells and transfer the content to the other falcon. Re-suspend.
9) Centrifuge 3000g for 10 minutes.
10) Repeat step 8 and 9 (in only one falcon) with 10 mL, 5 mL, 2.5 mL and finally add 0.625 mL (1/80 of initial volume).
11) During the centrifugation time, prepare the poly... tubes (label them) with recovery medium in them and put the cuvettes on ice: 1 of each at least has to be a control of cells without DNA, then 1 for each transformant you wish to make.
12) Transfer in a cuvette: 60 μL of cells + 1 μL of DNA (50ng/μL; none if control).
13) Pulse the cuvette.
14) Transfer immediately the content into the poly... tube (STERILE CONDITIONS).
15) Repeat 13, 14 and 15 for the number of prepared cuvettes.
16) Incubate the poly... tubes at 37°C for 3 to 6 hours.
17) Prepare plates with antibiotics (none for the control).
18) Note: ≈ 25 ml of LBA per petri dish, make sure the antibiotic is well diluted, labeling should be obvious.
19) Plate max 150 μL of transformed cells per petri dish and let grow overnight.
 "
"