Team:EPF-Lausanne/Our Project/Summary
From 2011.igem.org
(→Lysis-based transcription factor selection) |
|||
| Line 9: | Line 9: | ||
We ran platereader experiments involving growing cells to stationary growth phase and then induced with IPTG. The resulting optical density indicates that lysis occurs, with increased lysing efficiency as a function of increased IPTG concentration. | We ran platereader experiments involving growing cells to stationary growth phase and then induced with IPTG. The resulting optical density indicates that lysis occurs, with increased lysing efficiency as a function of increased IPTG concentration. | ||
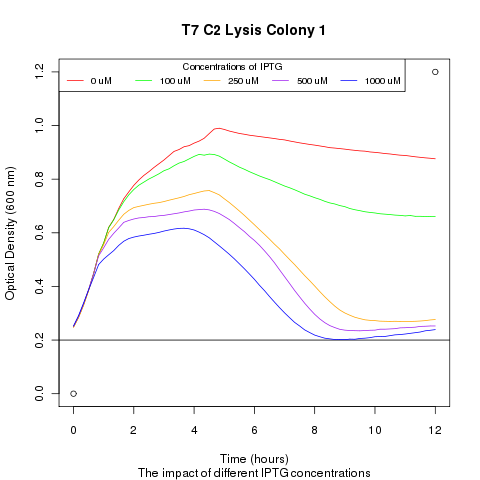
| - | + | [[File:t7_c2_lysis_col1.png|400px]] | |
''' 2) demonstrating that DNA could be recovered from the supernatant ''' | ''' 2) demonstrating that DNA could be recovered from the supernatant ''' | ||
| Line 15: | Line 15: | ||
We made a large culture of cells containing the lysis cassette and an RFP-containing plasmid and induced with IPTG. We collected samples of the supernatant every hour and quantified the amount of DNA in those samples by transforming the DNA and counting CFUs and by qPCR. Both quantifications revealed the same result: RFP plasmids were recovered in increasing amounts as time went on. | We made a large culture of cells containing the lysis cassette and an RFP-containing plasmid and induced with IPTG. We collected samples of the supernatant every hour and quantified the amount of DNA in those samples by transforming the DNA and counting CFUs and by qPCR. Both quantifications revealed the same result: RFP plasmids were recovered in increasing amounts as time went on. | ||
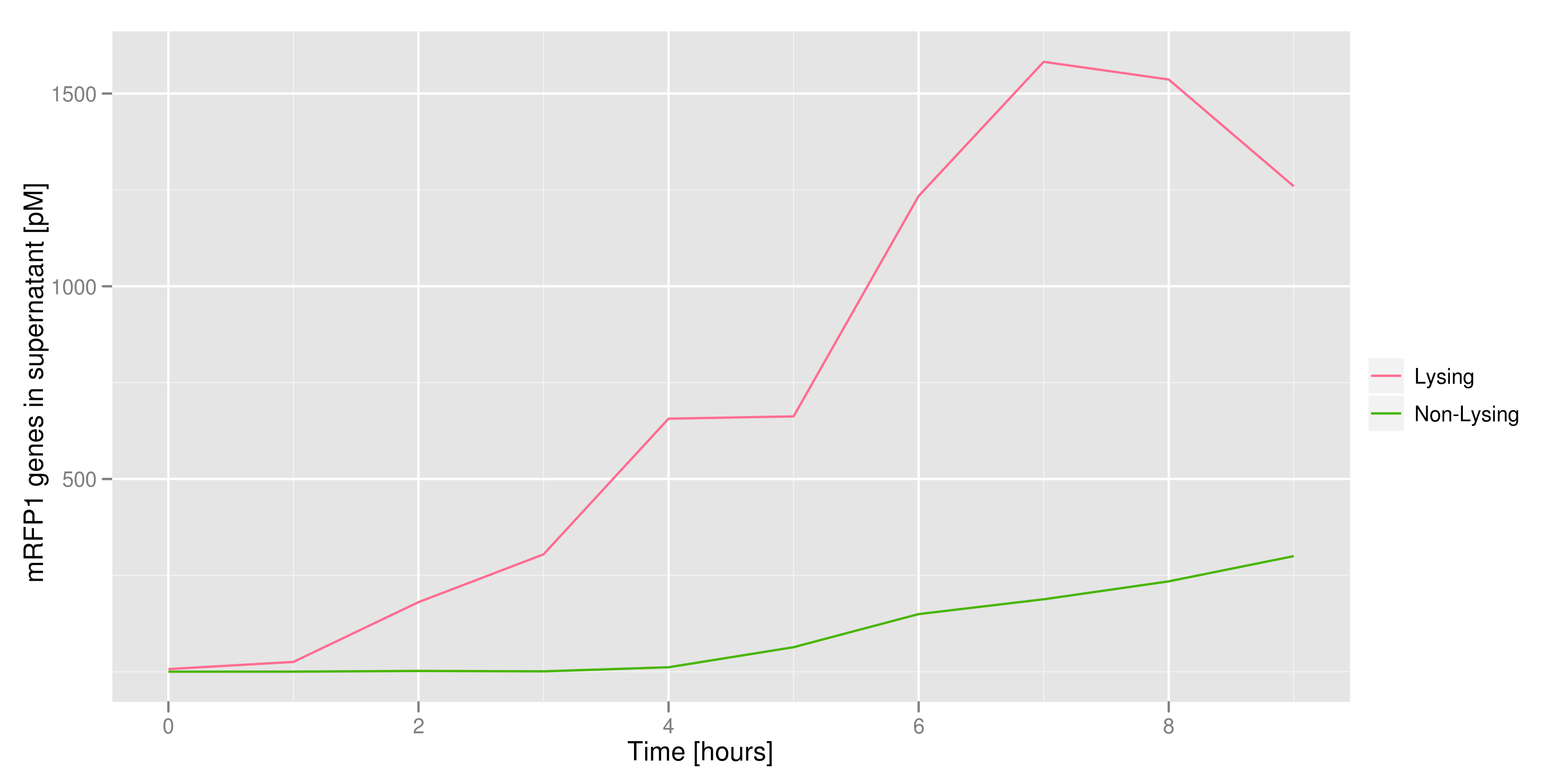
| - | + | [[File:First_DNA_supernatant.png|400px]] | |
''' 3) demonstrating that the appropriate cells (those with the desired trait or mutant DNA) would lyse, leaving the other cells intact ''' | ''' 3) demonstrating that the appropriate cells (those with the desired trait or mutant DNA) would lyse, leaving the other cells intact ''' | ||
| Line 38: | Line 38: | ||
''' 2) determining the binding energy landscape of each mutants | ''' 2) determining the binding energy landscape of each mutants | ||
| - | This is the ''in vitro'' characterization, which was performed thank to a microfluidic chip. This technique, called MITOMI, allows parallel testing of one mutant with | + | This is the ''in vitro'' characterization, which was performed thank to a microfluidic chip. This technique, called MITOMI, allows parallel testing of one mutant with 756 different DNA sequences. The absolute binding energy is then measured, and an enoLOGO is generated. |
| - | + | ||
''' 3) determining the binding of each mutant to the wild-type TetO sequence ''in vivo'' | ''' 3) determining the binding of each mutant to the wild-type TetO sequence ''in vivo'' | ||
| Line 50: | Line 49: | ||
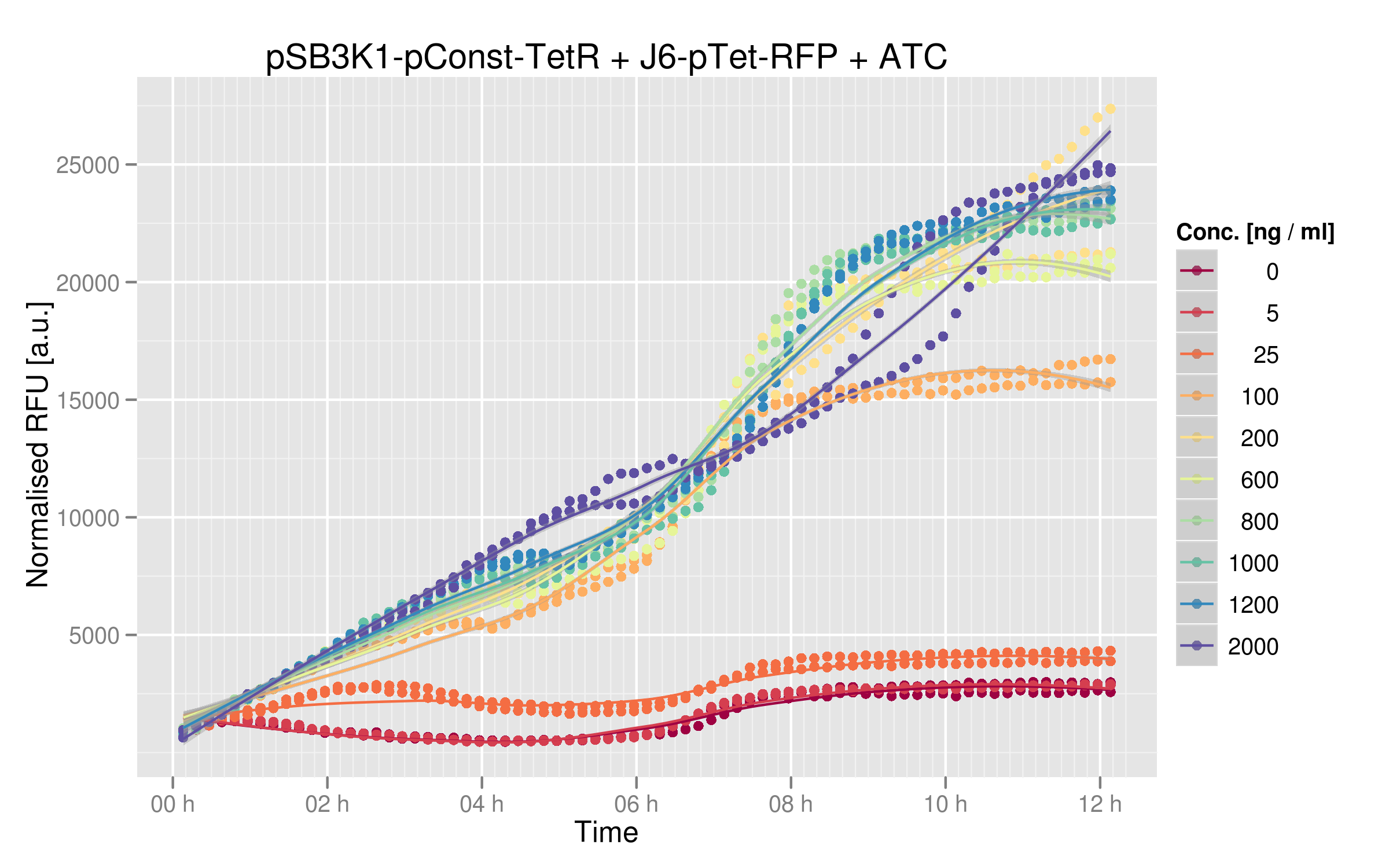
We ran platereader experiments for 6 of our mutants. Increasing concentrations of ATC were added to force expression of RFP, which allowed us to appreciate the repressive effect of TetR on fluorescence expression. The following graph shows wild-type TetR characterization: | We ran platereader experiments for 6 of our mutants. Increasing concentrations of ATC were added to force expression of RFP, which allowed us to appreciate the repressive effect of TetR on fluorescence expression. The following graph shows wild-type TetR characterization: | ||
| - | [[File:EPFL_Nadine-exp3-induction.png| | + | [[File:EPFL_Nadine-exp3-induction.png|400px]] |
| Line 57: | Line 56: | ||
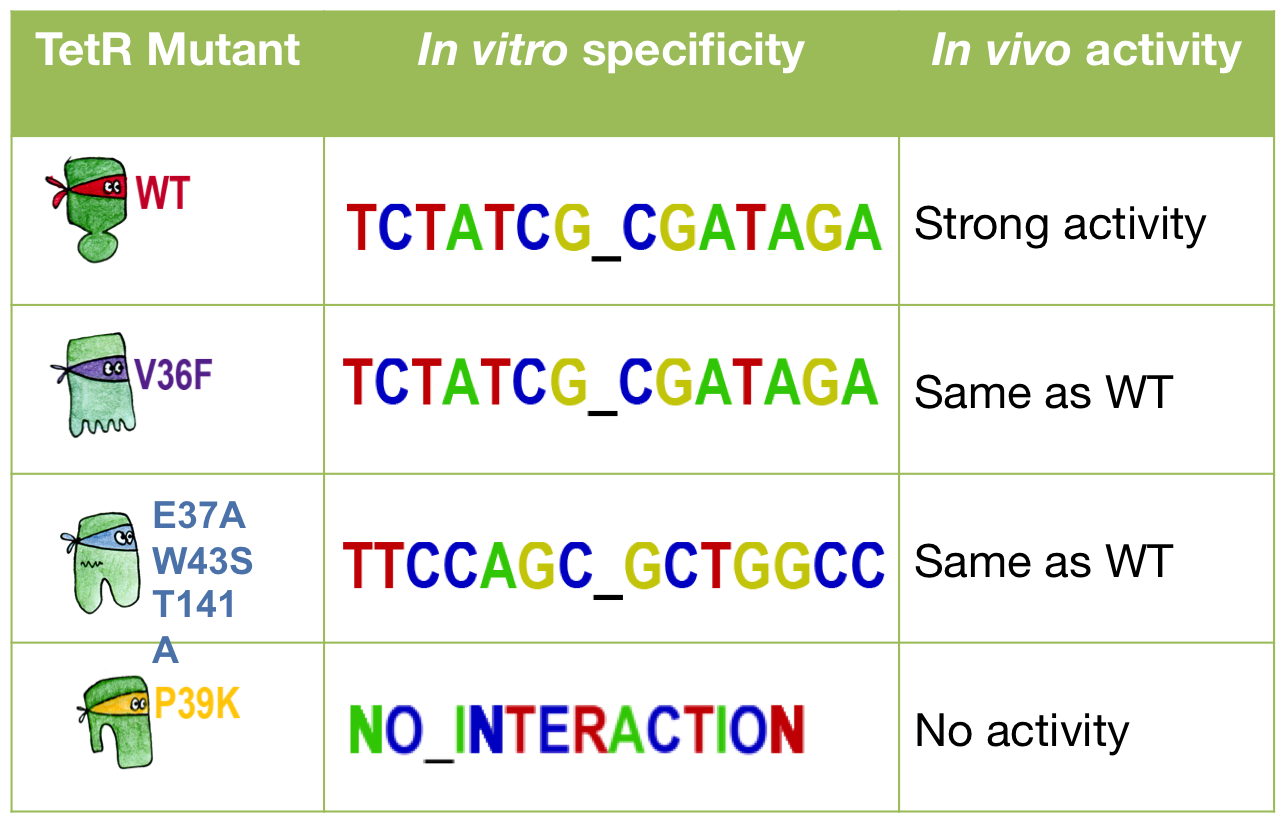
Three of our mutants plus the wild-type were tested both ''in vivo'' and ''in vitro''. The results were consistent between the two experiments. The V36F exhibits the same behaviour as the wild-type while the P39K shows no interaction to the DNA at all. The E37A triple mutant, however, has contradictory results: the MITOMI data indicate that this mutant has a change in specificity, with a consensus sequence being different from the wild-type Ptet. However, it still recognizes Ptet well, showing that there is still crosstalk with the endogenous promoter. | Three of our mutants plus the wild-type were tested both ''in vivo'' and ''in vitro''. The results were consistent between the two experiments. The V36F exhibits the same behaviour as the wild-type while the P39K shows no interaction to the DNA at all. The E37A triple mutant, however, has contradictory results: the MITOMI data indicate that this mutant has a change in specificity, with a consensus sequence being different from the wild-type Ptet. However, it still recognizes Ptet well, showing that there is still crosstalk with the endogenous promoter. | ||
| - | [[File:EPFL_Comparison_char.png| | + | [[File:EPFL_Comparison_char.png|500px]] |
{{:Team:EPF-Lausanne/Templates/Footer}} | {{:Team:EPF-Lausanne/Templates/Footer}} | ||
Revision as of 19:48, 28 October 2011
Results Summary
Lysis-based transcription factor selection
We implemented a prototype of our lysis selection system by experimentally validating four essential aspects of the strategy:
1) demonstrating that we could effectively lyse cells:
We ran platereader experiments involving growing cells to stationary growth phase and then induced with IPTG. The resulting optical density indicates that lysis occurs, with increased lysing efficiency as a function of increased IPTG concentration.
2) demonstrating that DNA could be recovered from the supernatant
We made a large culture of cells containing the lysis cassette and an RFP-containing plasmid and induced with IPTG. We collected samples of the supernatant every hour and quantified the amount of DNA in those samples by transforming the DNA and counting CFUs and by qPCR. Both quantifications revealed the same result: RFP plasmids were recovered in increasing amounts as time went on.
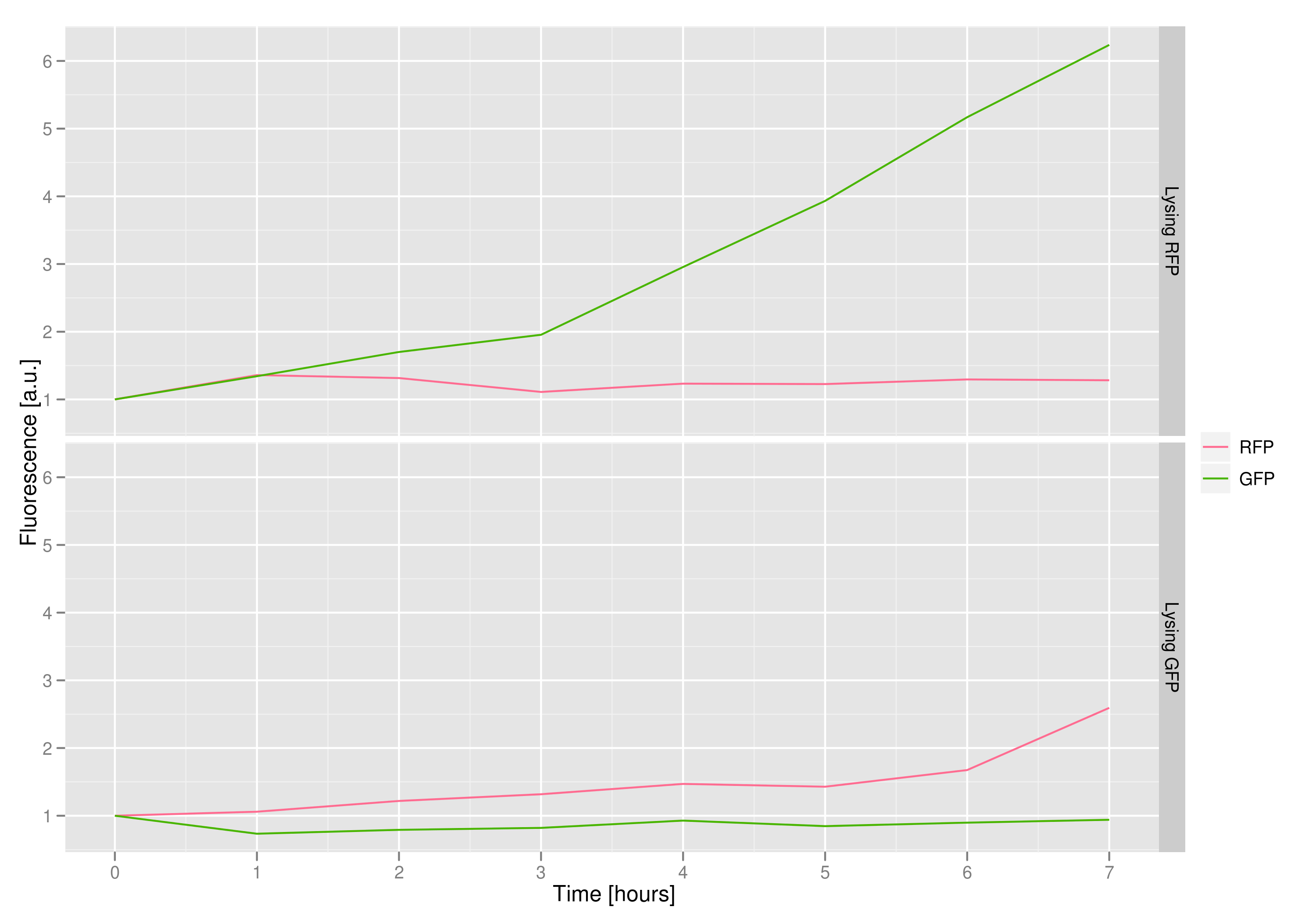
3) demonstrating that the appropriate cells (those with the desired trait or mutant DNA) would lyse, leaving the other cells intact
We made a co-culture of cells. One culture had the lysis cassette and an RFP plasmid, while the other had a GFP plasmid and no lysis cassette. Upon induction with IPTG, the qPCR and transformation methods of the previous experiment showed that RFP was recovered in large quantities and only trace quantities of GFP are detected.
4) demonstrating that promoter strength (i.e. transcription factor- DNA binding) has a direct impact on the amount and speed of lysing
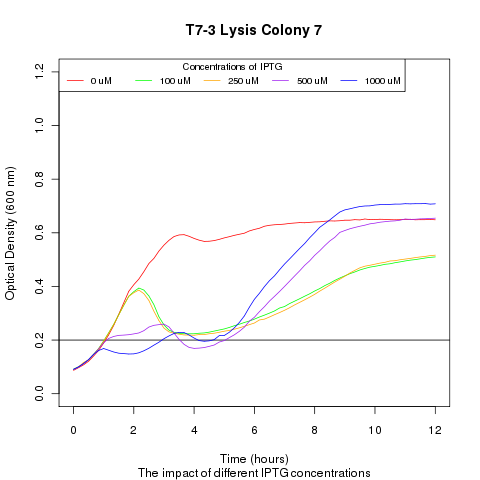
We ran a platereader experiment with multiple cultures, each culture containing a plasmid with a different T7 promoter mutant driving the lysis cassette. Induction with IPTG shows that having different promoters results in varying lysis strengths and speeds. This approach with promoter mutants supplies an alternate way of showing that favorable transcription-factor mutants will lyse cells more rapidly and efficiently then their peers. The resulting supernatant will therefore contain a proportionally high number of the better TF mutant DNA, which can be recovered and sequenced.
Characterisation
We decided to work with TetR, as a proof-of concept trascription factor characterization. We combined two approaches for characterizing our mutants, both in vitro and in vivo. We started by testing the wild-type and went on with some of our 12 TetR mutants. The strategy we used consisted of:
1) producing interesting TetR mutants
The mutants were created by site-specific and PCR-induced mutagenesis; we introduced point mutations at key amino acids involved in DNA recognition, in an attempt to alter the specificity of the TetR mutants.
2) determining the binding energy landscape of each mutants
This is the in vitro characterization, which was performed thank to a microfluidic chip. This technique, called MITOMI, allows parallel testing of one mutant with 756 different DNA sequences. The absolute binding energy is then measured, and an enoLOGO is generated.
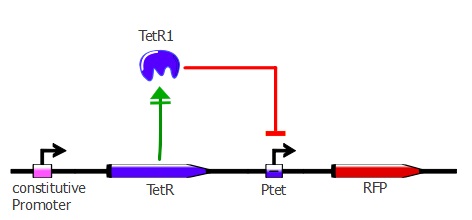
3) determining the binding of each mutant to the wild-type TetO sequence in vivo
For this characterization step, we set up a reporter construct: TetR constitutively expressed and RFP with a wild-type Ptet promoter, as you can see on the illustration below. The more RFP is expressed in the cells, the less the TetR mutant recognizes Ptet.
We ran platereader experiments for 6 of our mutants. Increasing concentrations of ATC were added to force expression of RFP, which allowed us to appreciate the repressive effect of TetR on fluorescence expression. The following graph shows wild-type TetR characterization:
4) comparing the results of both characterization
Three of our mutants plus the wild-type were tested both in vivo and in vitro. The results were consistent between the two experiments. The V36F exhibits the same behaviour as the wild-type while the P39K shows no interaction to the DNA at all. The E37A triple mutant, however, has contradictory results: the MITOMI data indicate that this mutant has a change in specificity, with a consensus sequence being different from the wild-type Ptet. However, it still recognizes Ptet well, showing that there is still crosstalk with the endogenous promoter.
 "
"