Team:Paris Bettencourt/Experiments/SinOp
From 2011.igem.org
| Line 156: | Line 156: | ||
</div> | </div> | ||
| - | <div id="scroll_left"><a href="https://2011.igem.org/Team:Paris_Bettencourt/ | + | <div id="scroll_left"><a href="https://2011.igem.org/Team:Paris_Bettencourt/SinOp"><img src="https://static.igem.org/mediawiki/2011/0/0a/Arrow-left-big.png" style="width:100%;"></a><a href="https://2011.igem.org/Team:Paris_Bettencourt/SinOp">SinOp design</a></div> |
</html> | </html> | ||
Revision as of 16:40, 27 October 2011

SinOp system experiments
Abstract
Results for the SinOp system:
- We successfully BioBricked both the KinA (BBa_K606046) constructs and sent them to the registry
Design overview

SinOp system
More information on the design here.
Parts and biobrick system construction
CLONING PLAN HERE
We suceeded in recovering the KinA gene from the non biobricked plasmid synthetized de novo by the 2009 Newcastle team, and cloned it into a standard biobrick plasmid, pSB1C3. Then we cloned this gene in front of the pVeg-SpovG (K143051) promoter + RBS. These two constructs had bees sended to the registry into pSB1C3.
This construct has been cloned right away into a replicative plasmid for subtilis and transformed, and we are caracterizing it at the moment.
Characterization of the sporulation induced by the expression of KinA
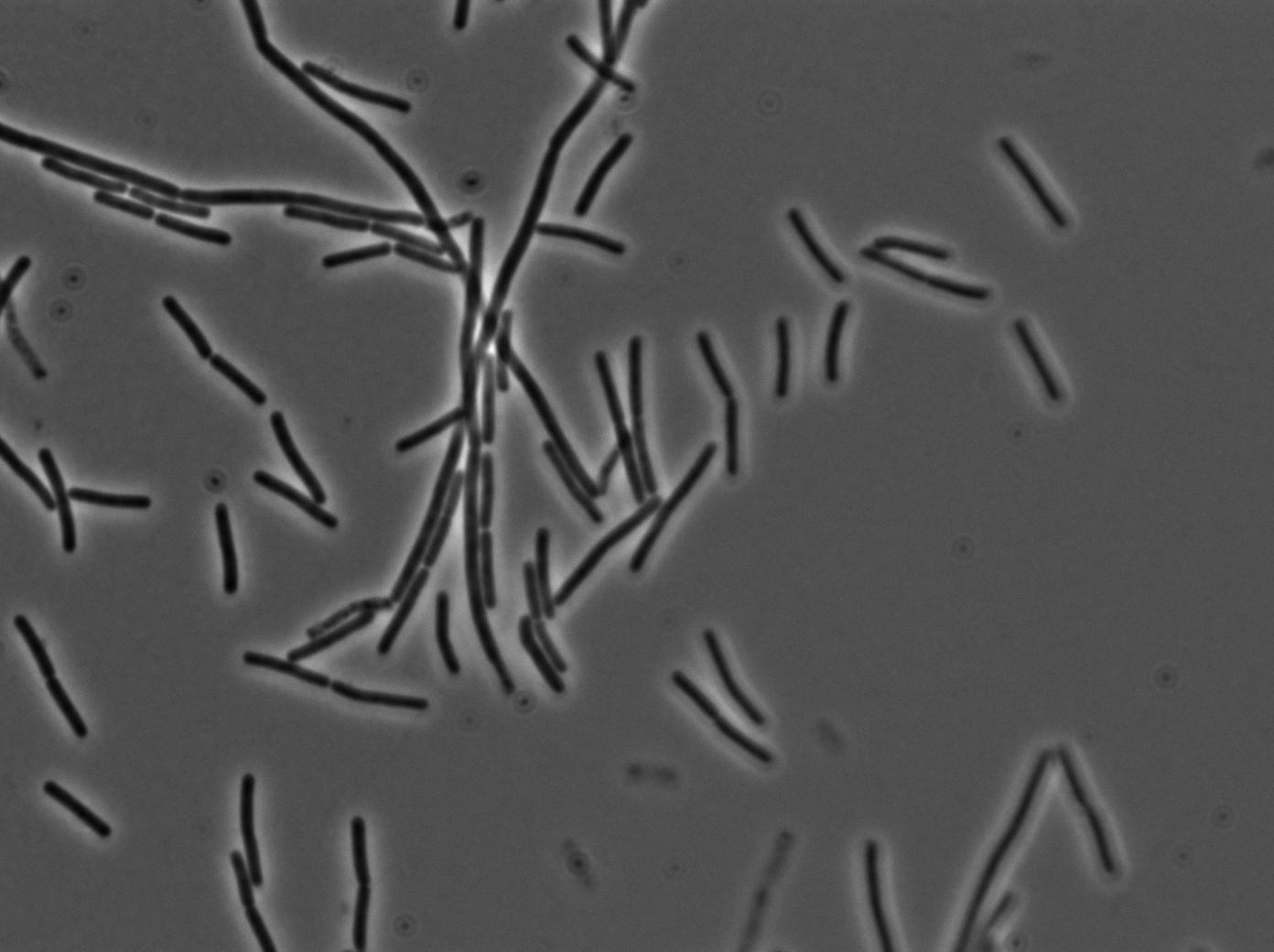
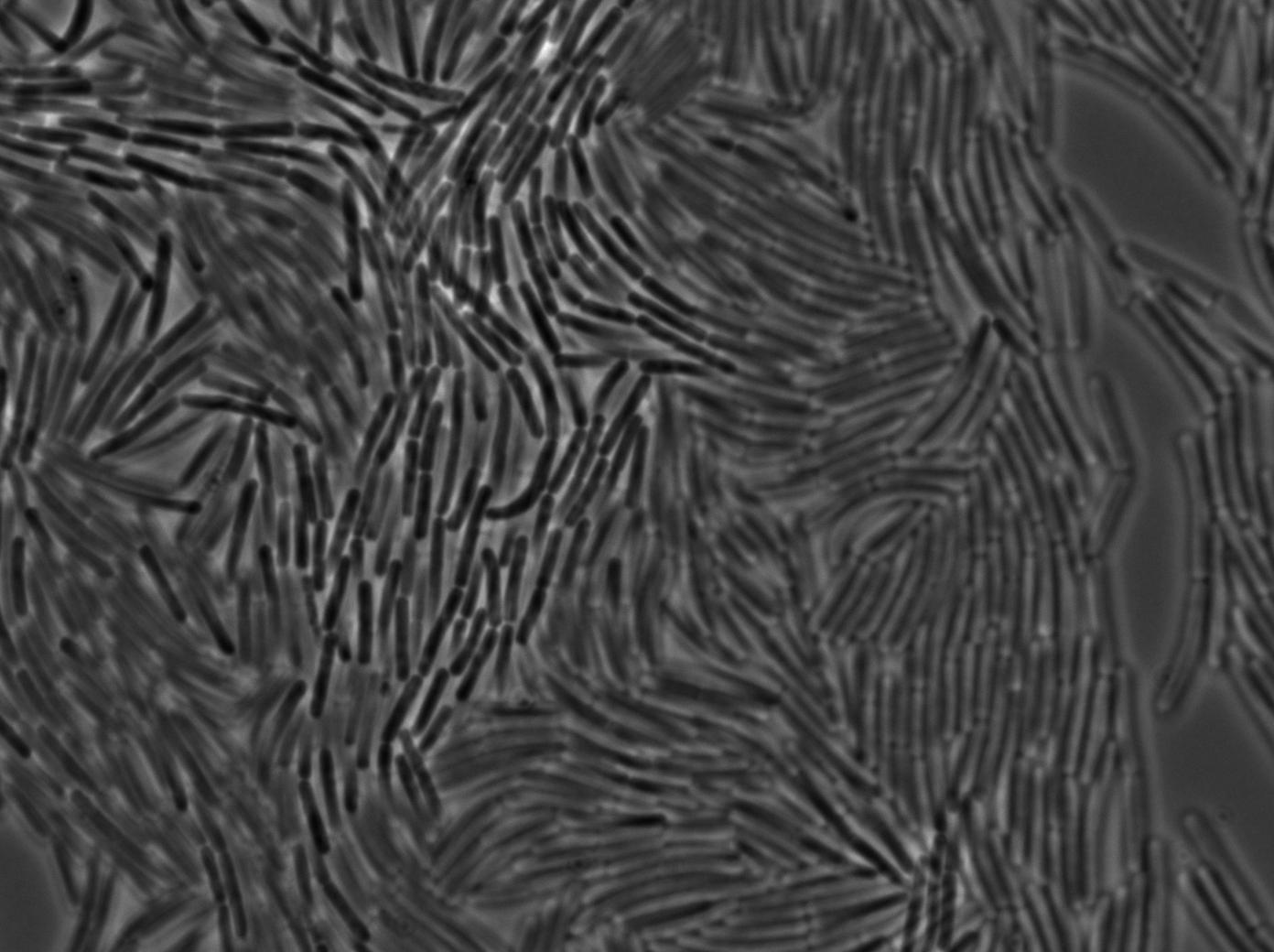
We used a strain holding a KinA gene under the control of a hyperspank promoter. Growing the cell into a synthetic minimal media for for hours, we saw the cell starting soprulating under the microscope after 1h30. Here are the images from the characterization.
Ref: Single,chemically defined sporulation medium for bacillus subtilis: growth,sporulation,and extracellular protease production. James H.HAGEMAN et al
We suceeded in recovering the KinA gene from the non biobricked plasmid synthetized de novo by the 2009 Newcastle team, and cloned it into a standard biobrick plasmid, pSB1C3. Then we cloned this gene in front of the pVeg-SpovG (K143051) promoter + RBS. These two constructs had bees sended to the registry into pSB1C3.
This construct has been cloned right away into a replicative plasmid for subtilis and transformed, and we are caracterizing it at the moment.
Characterizing the SinI system
As explained in the design page
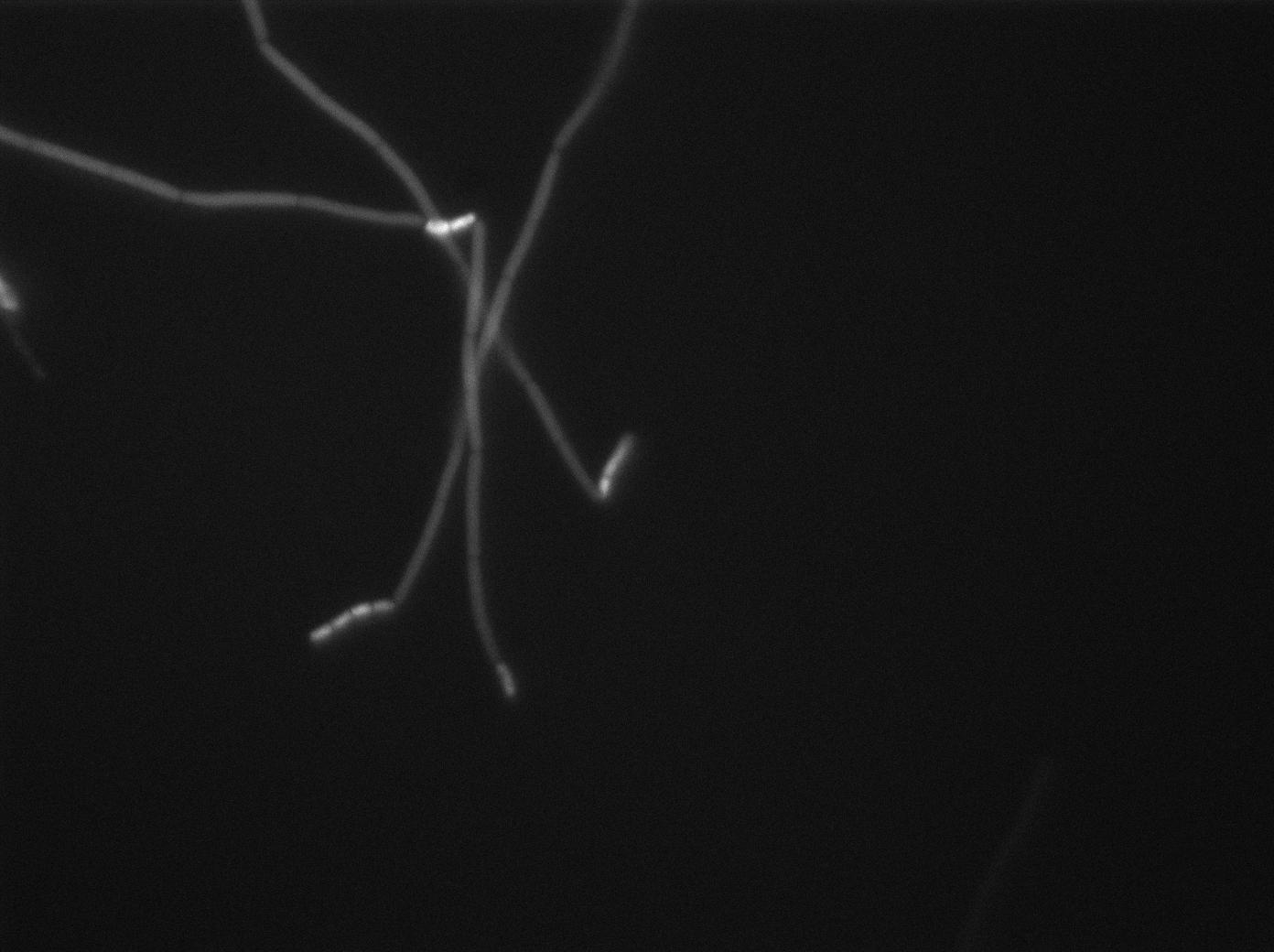
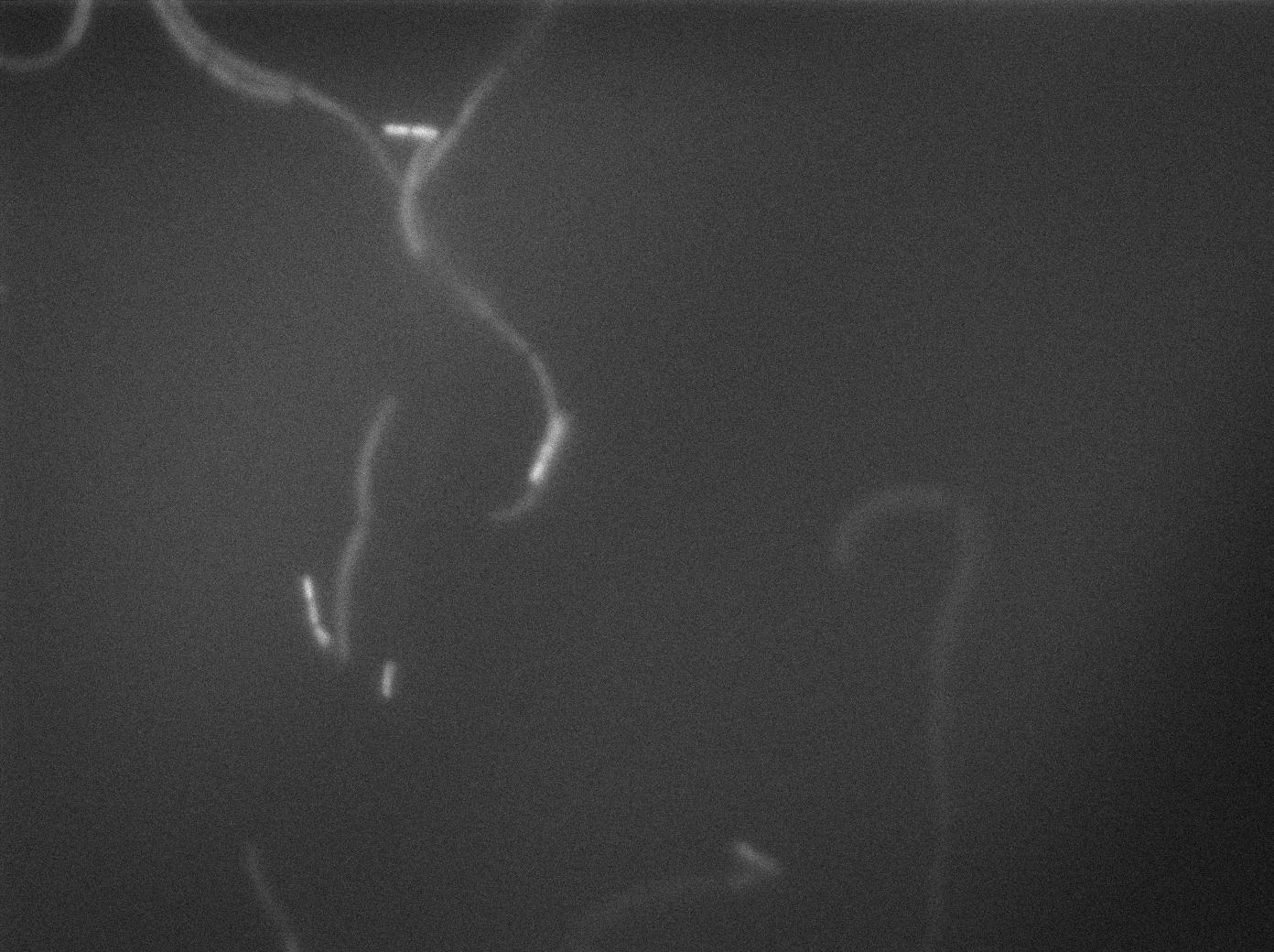
In order to verify the function of SinI, we tested its activity under the Hyperspank promoter. Upon induction, a clear phenotypic change occured where cells turned into chain-like growth regime (fluorescent cellsin image), unlike wild-type cells that kept their regular elongated separated cell phenotype (non-fluorecent cells in image).
You can find the video of our results for the SinI system with IPTG induction here and without IPTG here.
Preparation of slides
Dilution of overnight cultures : YC164 and YC227 (SinOp Design) .
YC227 produces SinI which enhances biofilm formation. If SinI diffuses through the nanotubes we expect to see fluorescence from Peps-gfp construct in the receiver cells.
Two well slides : 1-control (YC164 with IPTG) 2-Mix (both strains with IPTG)
Observation
-37°C Microscopy-
We observed the plate with TRANS and YFP-filter settings on the Old Zeiss. Unfortunately, this microscope gets quickly out of focus so we need to take each picture manually. As expected, YC227 was producing GFP at the begining of the experiment, while YC164 was not. Our observations over 4 hours are the following:
- YC164 grows fast (several divisions over the experiments)
- YC227 does not grow at all and keeps its fluorescence pretty well
- YC164 does not change its state (no obvious biofilm formation)
- After letting the plate overnight we still see some florescence but not as if YC164 changed its fluorescence status.
Our conclusions
 "
"