Team:EPF-Lausanne/Our Project/Reporter Systems/tetR
From 2011.igem.org
(→TetR mutant characterization) |
|||
| (17 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
=== Description === | === Description === | ||
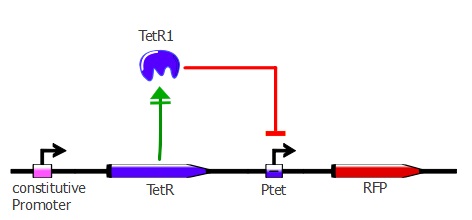
| - | + | This first readout system is the simplest one; it is composed of TetR driven by a constitutive promoter with RFP (red fluorescent protein) under Ptet control. If TetR '''binds''' to Ptet, then RFP is ''' repressed'''. By applying different concentrations of ATC (anhydrotetracycline) to the cells, RFP is expressed - the stronger the TetR mutant binds to Ptet, the higher the ATC concentration needed to have full expression of RFP. The genetic cascade is depicted in the illustration below. | |
[[File:EPFL_Summary_without_LacI.jpg]] | [[File:EPFL_Summary_without_LacI.jpg]] | ||
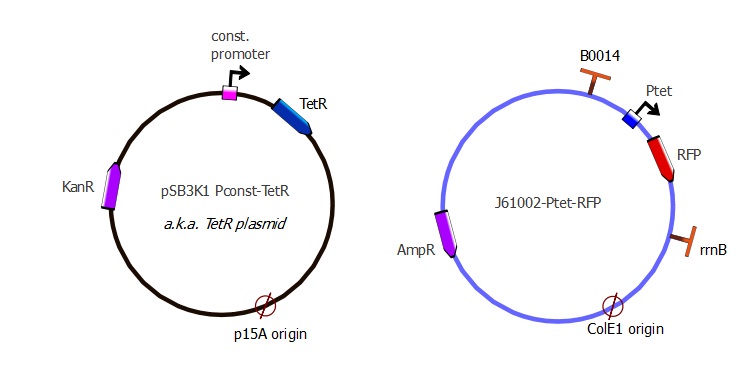
| - | The TetR and RFP genes were put on two different plasmids: pSB3K1 pConst-TetR and J61002 Ptet-RFP. For more details about them, please refer to the [https://2011.igem.org/Team:EPF-Lausanne/Our_Project/ | + | The TetR and RFP genes were put on two different plasmids: pSB3K1 pConst-TetR and J61002 Ptet-RFP. For more details about them, please refer to the [https://2011.igem.org/Team:EPF-Lausanne/Our_Project/Reporter_Systems/plasmid Plasmids details] page. |
[[File:EPFL_TetR_and_Ptet-RFP.jpg|500px]] | [[File:EPFL_TetR_and_Ptet-RFP.jpg|500px]] | ||
| Line 18: | Line 18: | ||
| - | ''ATC induction'' | + | ====''ATC induction''==== |
| - | To validate our first readout system, we performed platereader experiments with different concentrations of anhydrotetracycline (ATC). This molecule binds to the TetR dimers and induces conformational changes that make the transcription factor unable to bind to Ptet. As a result, RFP can be expressed | + | To validate our first readout system, we performed platereader experiments with different concentrations of anhydrotetracycline (ATC). This molecule binds to the TetR dimers and induces conformational changes that make the transcription factor unable to bind to Ptet. As a result, RFP can be expressed in the cell, as shown in the cartoons below: |
Gene expression in the cell without ATC: | Gene expression in the cell without ATC: | ||
| Line 30: | Line 30: | ||
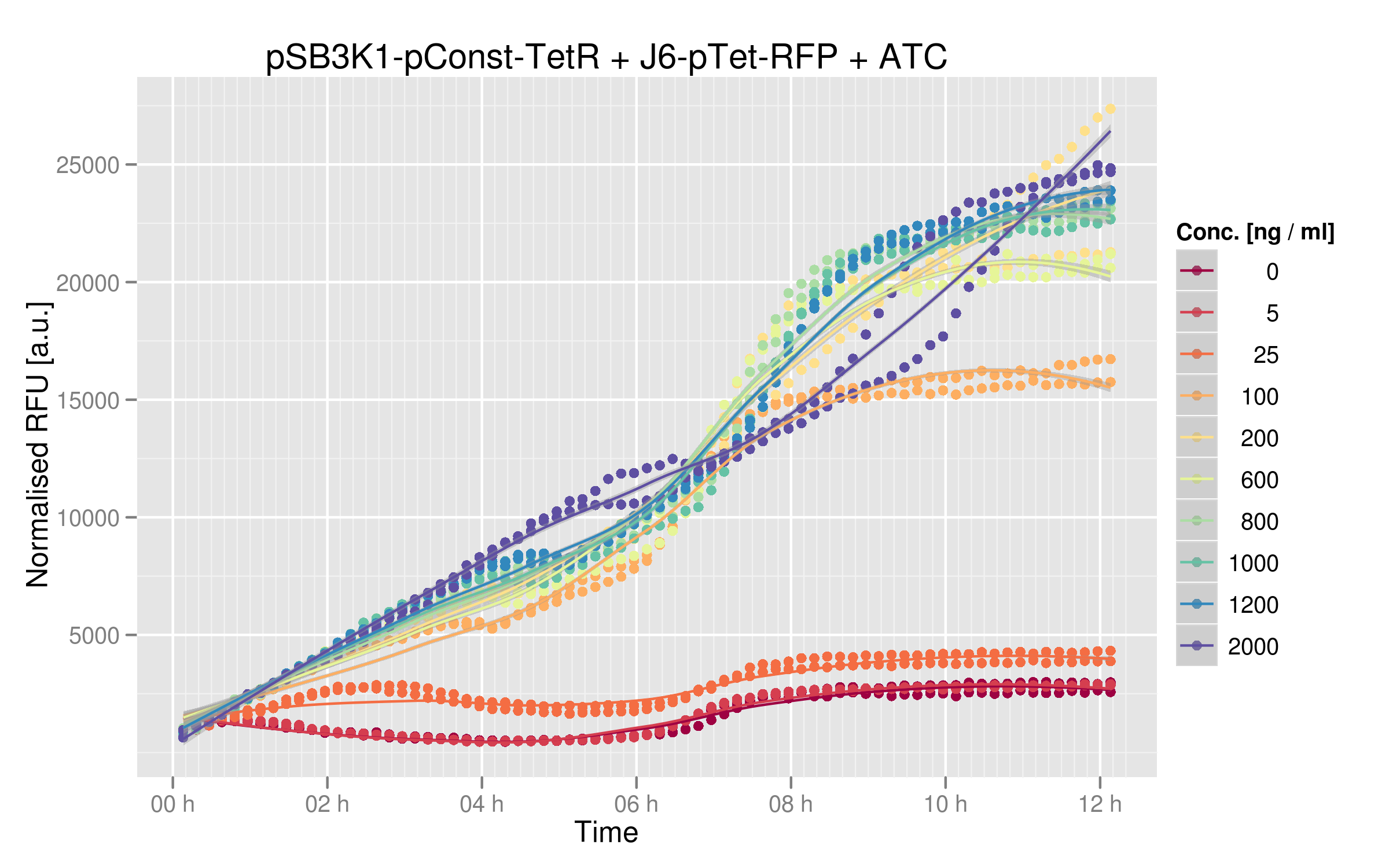
| - | Here are the results of our ATC induction experiment: | + | Here are the results of our ATC induction experiment, performed on the wild-type TetR: |
[[File:EPFL_Nadine-exp3-induction.png|600px]] | [[File:EPFL_Nadine-exp3-induction.png|600px]] | ||
| - | The induction curves show that RFP expression increases with addition of ATC. In the absence of ATC, cells express about 2500 normalized RFUs (relative fluorescence units) when they reach a plateau, whereas at the highest concentrations of ATC we observe 25'000 normalized RFUs. There is a 10x difference between normal medium (without ATC) and addition of ATC, showing that our first readout system is sensitive to ATC concentration. Moreover, expression of RFP in cells without ATC is 10-fold lower than the values obtained for [https://2011.igem.org/ | + | The induction curves show that RFP expression increases with addition of ATC. In the absence of ATC, cells express about 2500 normalized RFUs (relative fluorescence units) when they reach a plateau, whereas at the highest concentrations of ATC we observe 25'000 normalized RFUs. There is a 10x difference between normal medium (without ATC) and addition of ATC, showing that our first readout system is sensitive to ATC concentration. Moreover, expression of RFP in cells without ATC is 10-fold lower than the values obtained for [https://2011.igem.org/Team:EPF-Lausanne/Our_Project/Reporter_Systems/ptet Ptet characterization] alone. |
| - | We can reasonably assume that this system can be used for in vivo screening: TetR mutants that are not capable of recognizing the consensus Ptet sequence will yield more RFP than other mutants | + | We can reasonably assume that this system can be used for in vivo screening: TetR mutants that are not capable of recognizing the consensus Ptet sequence will yield more RFP than other mutants still recognizing Ptet. The 10x difference between RFU values in this experiment seems to be a useful window for characterizing our mutants. |
| - | ''Dose-response'' | + | ====''Dose-response''==== |
[[File:EPFL_Nadine-exp3-doseresponse.png|600px]] | [[File:EPFL_Nadine-exp3-doseresponse.png|600px]] | ||
| - | By looking at the dose-response graph, we can see a significant increase between 0 and 200 ng/microL ATC; then the RFU values begin to plateau. The graph shows that the TetR-Ptet interaction has a strong impact on RFP expression. The highest RFUs measured here correspond to the levels of the Ptet-RFP construct alone (in the absence of the the TetR plasmid), | + | By looking at the dose-response graph, we can see a significant increase between 0 and 200 ng/microL ATC; then the RFU values begin to plateau. The graph shows that the TetR-Ptet interaction has a strong impact on RFP expression. The highest RFUs measured here correspond to the levels of the Ptet-RFP construct alone (in the absence of the the TetR plasmid, see [https://2011.igem.org/Team:EPF-Lausanne/Our_Project/Reporter_Systems/ptet Ptet characterization]), demonstrating complete TetR suppression in our experiments. |
| + | |||
| Line 51: | Line 52: | ||
| - | With the wild-type TetR in our system, RFP is repressed because wt-TetR binds to Ptet. However, mutants can potentially recognize a different promoter sequence | + | With the wild-type TetR in our system, RFP is repressed because wt-TetR binds to Ptet with a great efficiency. However, mutants can potentially recognize a different promoter sequence than Ptet and thus would be unable to repress RFP expression, as seen in the cartoons: |
Gene expression with wild-type TetR: | Gene expression with wild-type TetR: | ||
| Line 61: | Line 62: | ||
| - | We were able to test 6 of our TetR mutants with this first reporter system. For each of them, we | + | We were able to test 6 of our TetR mutants with this first reporter system. For each of them, we inserted the mutated TetR sequence in the [https://2011.igem.org/Team:EPF-Lausanne/Our_Project/Reporter_Systems/plasmid#First_plasmid_-_pSB3K1_Pconst-TetR Pconst-TetR plasmid] instead of the wild-type TetR. |
| - | '' | + | ====''Mutants individual results''==== |
For each TetR mutant tested, the individual ''in vivo'' characterization results are displayed on their respective Registry page: | For each TetR mutant tested, the individual ''in vivo'' characterization results are displayed on their respective Registry page: | ||
| Line 75: | Line 76: | ||
| - | ''Mutants comparison'' | + | ====''Mutants comparison''==== |
In order to see better the differences between different mutants, we made two graphs: one showing the induction curves of all six mutants without ATC and one showing all six mutants with the maximal ATC concentration (2000 ng/mL). | In order to see better the differences between different mutants, we made two graphs: one showing the induction curves of all six mutants without ATC and one showing all six mutants with the maximal ATC concentration (2000 ng/mL). | ||
| Line 86: | Line 87: | ||
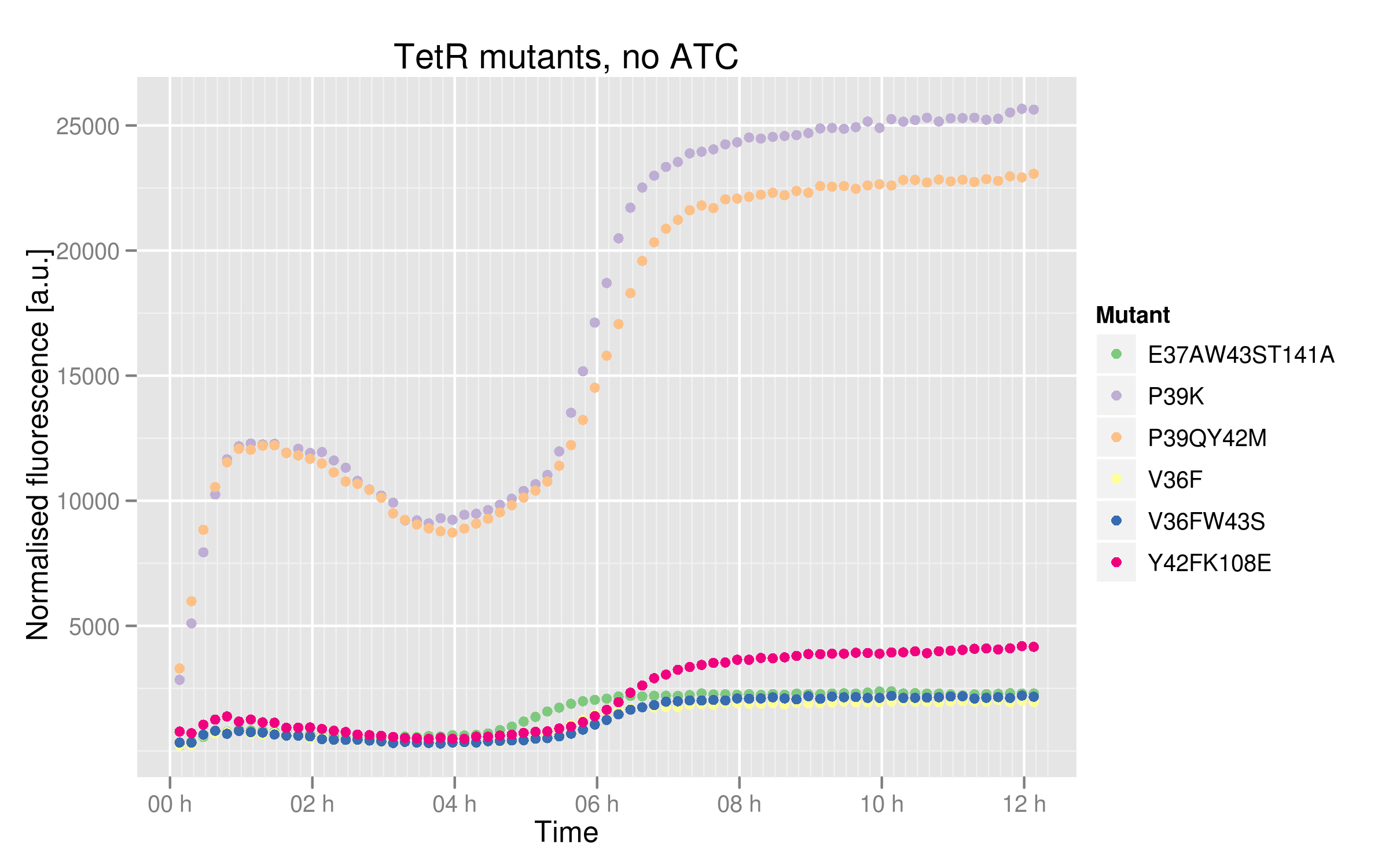
* V36F, V36F W43S and E37A W43S T141A mutants plateau near 2500 normalized RFUs, which is the value observer for the wild-type without ATC. This means that these mutants can recognize and repress Ptet with the same strength as the wild-type. | * V36F, V36F W43S and E37A W43S T141A mutants plateau near 2500 normalized RFUs, which is the value observer for the wild-type without ATC. This means that these mutants can recognize and repress Ptet with the same strength as the wild-type. | ||
* Y42F K108E mutant reaches about 5000 normalized RFUs; this value is quite low compared to Ptet intrinsic strength but it is twice higher than RFP expression induced by the wild-type, indicating that this mutant recognizes and represses Ptet with less power that the wild-type | * Y42F K108E mutant reaches about 5000 normalized RFUs; this value is quite low compared to Ptet intrinsic strength but it is twice higher than RFP expression induced by the wild-type, indicating that this mutant recognizes and represses Ptet with less power that the wild-type | ||
| - | * P39K and P39Q Y42M mutants induce very high RFP expression, 25000 RFUs being the maximal fluorescence obtained by the promoter alone. These data | + | * P39K and P39Q Y42M mutants induce very high RFP expression, 25000 RFUs being the maximal fluorescence obtained by the promoter alone. These data then hint at the fact that the two mutants are not able to bind and repress Ptet. |
| Line 92: | Line 93: | ||
[[File:EPFL_TetR-ATC2000-induction.png|600px]] | [[File:EPFL_TetR-ATC2000-induction.png|600px]] | ||
| - | These data tell us that some mutants are not as much | + | These data tell us that some mutants are not as much affected by ATC as the others. P39K, P39Q Y42M and Y42F K108E mutants seem to be completely repressed, reaching the maximal RFUs of 25000 (based on Ptet characterization and wild-type TetR data). However, V36F, V36F W43S and E37A W43S T141A only plateau at 15000 RFUs. Still, their dose-response curves do indicate that we reached maximal TetR inhibition. A plausible explanation for this is that these 3 mutants cannot bind ATC properly anymore. There could be intrinsic differences in the cells used, such as endogenous TetR levels expression, but we used the same competent cell batch for all our mutant transformations. |
| + | |||
| + | |||
| + | ==== ''Summary of the results''==== | ||
| + | |||
| + | Our first reporter system allowed us to successfully characterize 6 of our mutants; these results are listed in the following table: | ||
| + | |||
| + | {| | ||
| + | !| Mutant | ||
| + | !| Biobrick number | ||
| + | !| Affinity compared to WT | ||
| + | !| ATC repression | ||
| + | |- | ||
| + | | V36F | ||
| + | | <html> <a href="http://partsregistry.org/Part:BBa_K613013"> BBa_K613013</a> </html> | ||
| + | | same | ||
| + | | altered | ||
| + | |- | ||
| + | | V36F W43S | ||
| + | | <html> <a href="http://partsregistry.org/Part:BBa_K613014"> BBa_K613014</a> </html> | ||
| + | | same | ||
| + | | altered | ||
| + | |- | ||
| + | | E37A W43S T141A | ||
| + | | <html> <a href="http://partsregistry.org/Part:BBa_K613015"> BBa_K613015</a> </html> | ||
| + | | same | ||
| + | | altered | ||
| + | |- | ||
| + | | P39K | ||
| + | | <html> <a href="http://partsregistry.org/Part:BBa_K613016"> BBa_K613016</a> </html> | ||
| + | | no affinity | ||
| + | | normal | ||
| + | |- | ||
| + | | Y42F K108E | ||
| + | | <html> <a href="http://partsregistry.org/Part:BBa_K613018"> BBa_K613018</a> </html> | ||
| + | | reduced | ||
| + | | normal | ||
| + | |- | ||
| + | | P39Q Y42M | ||
| + | | <html> <a href="http://partsregistry.org/Part:BBa_K613019"> BBa_K613019</a> </html> | ||
| + | | no affinity | ||
| + | | normal | ||
| + | |} | ||
| + | |||
| - | |||
| - | + | === Comparison with in vitro data === | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | A table summing up the results obtained for the TetR mutants, both ''in vitro'' and ''in vivo'', is available [https://2011.igem.org/Team:EPF-Lausanne/Our_Project/TetR_mutants/Conclusion here]. | ||
{{:Team:EPF-Lausanne/Templates/Footer}} | {{:Team:EPF-Lausanne/Templates/Footer}} | ||
Latest revision as of 19:36, 26 October 2011
In Vivo Characterization
In vivo Main | TetR-RFP System | TetR-LacI-RFP System | Ptet Characterization | Plac Characterization | Plasmid Details
Contents |
TetR - RFP system
Description
This first readout system is the simplest one; it is composed of TetR driven by a constitutive promoter with RFP (red fluorescent protein) under Ptet control. If TetR binds to Ptet, then RFP is repressed. By applying different concentrations of ATC (anhydrotetracycline) to the cells, RFP is expressed - the stronger the TetR mutant binds to Ptet, the higher the ATC concentration needed to have full expression of RFP. The genetic cascade is depicted in the illustration below.
The TetR and RFP genes were put on two different plasmids: pSB3K1 pConst-TetR and J61002 Ptet-RFP. For more details about them, please refer to the Plasmids details page.
Experimental validation - with wild-type TetR
ATC induction
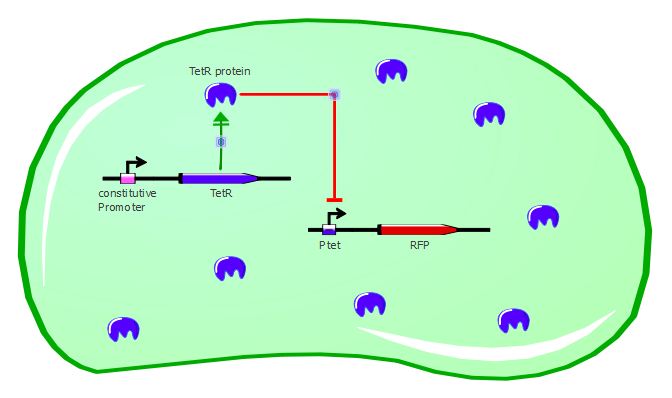
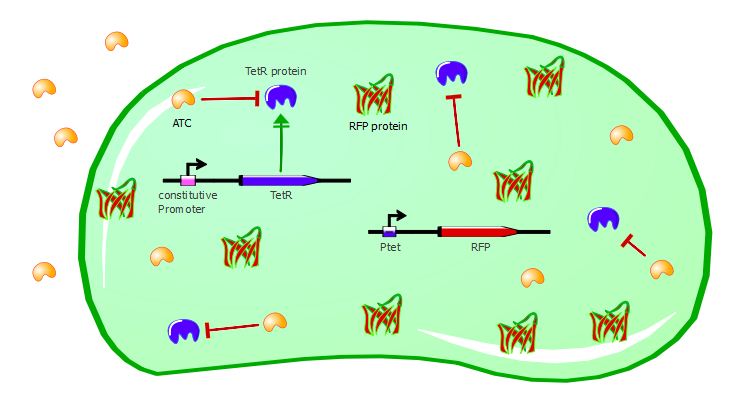
To validate our first readout system, we performed platereader experiments with different concentrations of anhydrotetracycline (ATC). This molecule binds to the TetR dimers and induces conformational changes that make the transcription factor unable to bind to Ptet. As a result, RFP can be expressed in the cell, as shown in the cartoons below:
Gene expression in the cell without ATC:
Gene expression in the cell with ATC:
Here are the results of our ATC induction experiment, performed on the wild-type TetR:
The induction curves show that RFP expression increases with addition of ATC. In the absence of ATC, cells express about 2500 normalized RFUs (relative fluorescence units) when they reach a plateau, whereas at the highest concentrations of ATC we observe 25'000 normalized RFUs. There is a 10x difference between normal medium (without ATC) and addition of ATC, showing that our first readout system is sensitive to ATC concentration. Moreover, expression of RFP in cells without ATC is 10-fold lower than the values obtained for Ptet characterization alone.
We can reasonably assume that this system can be used for in vivo screening: TetR mutants that are not capable of recognizing the consensus Ptet sequence will yield more RFP than other mutants still recognizing Ptet. The 10x difference between RFU values in this experiment seems to be a useful window for characterizing our mutants.
Dose-response
By looking at the dose-response graph, we can see a significant increase between 0 and 200 ng/microL ATC; then the RFU values begin to plateau. The graph shows that the TetR-Ptet interaction has a strong impact on RFP expression. The highest RFUs measured here correspond to the levels of the Ptet-RFP construct alone (in the absence of the the TetR plasmid, see Ptet characterization), demonstrating complete TetR suppression in our experiments.
TetR mutant characterization
With the wild-type TetR in our system, RFP is repressed because wt-TetR binds to Ptet with a great efficiency. However, mutants can potentially recognize a different promoter sequence than Ptet and thus would be unable to repress RFP expression, as seen in the cartoons:
Gene expression with wild-type TetR:

Gene expression with a TetR mutant not recognizing Ptet:
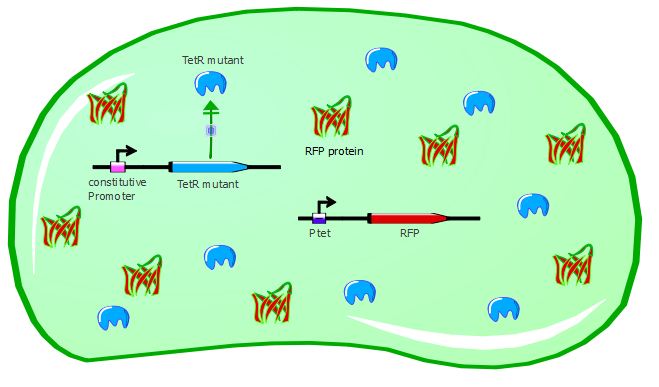
We were able to test 6 of our TetR mutants with this first reporter system. For each of them, we inserted the mutated TetR sequence in the Pconst-TetR plasmid instead of the wild-type TetR.
Mutants individual results
For each TetR mutant tested, the individual in vivo characterization results are displayed on their respective Registry page:
- V36F mutant: [http://partsregistry.org/Part:BBa_K613013 K613013]
- V36F W43S double mutant: [http://partsregistry.org/Part:BBa_K613014 K613014]
- E37A W43S T141A triple mutant: [http://partsregistry.org/Part:BBa_K613015 K613015]
- P39K mutant: [http://partsregistry.org/Part:BBa_K613016 K613016]
- Y42F K108E double mutant: [http://partsregistry.org/Part:BBa_K613018 K613018]
- P39Q Y42M double mutant: [http://partsregistry.org/Part:BBa_K613019 K613019]
Mutants comparison
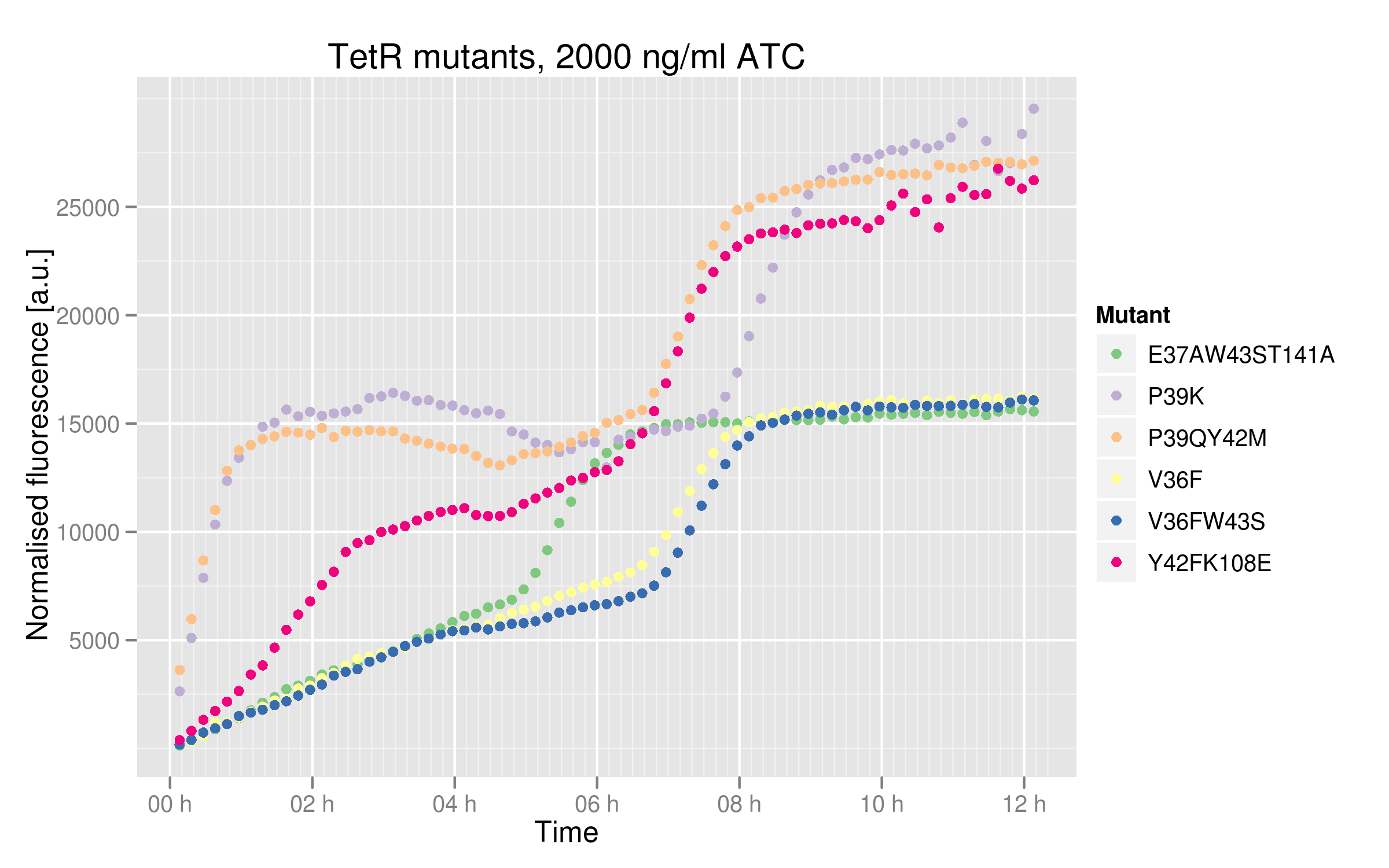
In order to see better the differences between different mutants, we made two graphs: one showing the induction curves of all six mutants without ATC and one showing all six mutants with the maximal ATC concentration (2000 ng/mL).
Based on this graph, we can estimate the binding affinity of the mutants to consensus Ptet:
- V36F, V36F W43S and E37A W43S T141A mutants plateau near 2500 normalized RFUs, which is the value observer for the wild-type without ATC. This means that these mutants can recognize and repress Ptet with the same strength as the wild-type.
- Y42F K108E mutant reaches about 5000 normalized RFUs; this value is quite low compared to Ptet intrinsic strength but it is twice higher than RFP expression induced by the wild-type, indicating that this mutant recognizes and represses Ptet with less power that the wild-type
- P39K and P39Q Y42M mutants induce very high RFP expression, 25000 RFUs being the maximal fluorescence obtained by the promoter alone. These data then hint at the fact that the two mutants are not able to bind and repress Ptet.
These data tell us that some mutants are not as much affected by ATC as the others. P39K, P39Q Y42M and Y42F K108E mutants seem to be completely repressed, reaching the maximal RFUs of 25000 (based on Ptet characterization and wild-type TetR data). However, V36F, V36F W43S and E37A W43S T141A only plateau at 15000 RFUs. Still, their dose-response curves do indicate that we reached maximal TetR inhibition. A plausible explanation for this is that these 3 mutants cannot bind ATC properly anymore. There could be intrinsic differences in the cells used, such as endogenous TetR levels expression, but we used the same competent cell batch for all our mutant transformations.
Summary of the results
Our first reporter system allowed us to successfully characterize 6 of our mutants; these results are listed in the following table:
| Mutant | Biobrick number | Affinity compared to WT | ATC repression |
|---|---|---|---|
| V36F | BBa_K613013 | same | altered |
| V36F W43S | BBa_K613014 | same | altered |
| E37A W43S T141A | BBa_K613015 | same | altered |
| P39K | BBa_K613016 | no affinity | normal |
| Y42F K108E | BBa_K613018 | reduced | normal |
| P39Q Y42M | BBa_K613019 | no affinity | normal |
Comparison with in vitro data
A table summing up the results obtained for the TetR mutants, both in vitro and in vivo, is available here.
 "
"