Team:Paris Bettencourt/tRNA diffusion
From 2011.igem.org
| (45 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
<h2>The amber codon and the tRNA amber suppressor</h2> | <h2>The amber codon and the tRNA amber suppressor</h2> | ||
| + | </td> | ||
| + | <td> </td> | ||
| + | <td> | ||
<table> | <table> | ||
<tr> | <tr> | ||
<td VALIGN=CENTER> | <td VALIGN=CENTER> | ||
| - | <p>The <em>amber codon</em> is one of the less used stop codon in bacteria. The principle of the artificial amber | + | <p>The <em>amber codon</em> is one of the less used stop codon in bacteria. The principle of the artificial amber suppressor tRNA is to provide a tRNA corresponding to this stop codon.</p> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | <p>When the ribosome translates the RNA into a protein, it first looks for the RBS sequence and fixes on it. It tries to fit the codon it is located on with the three complementary bases of tRNA flying round. When it finds the correct tRNA with the <em>anti-codon</em> of the start codon with a methionine loaded on it, the translation starts. Then, codon after codon, the ribosome tries to fit many tRNA on the codon it is placed on, until it find the correct one, fixes the corresponding amino-acid and then moves to the next codon. When the ribosome reaches a codon that is sotp codon in the genetic code of the cell, the translation stops.</p> | ||
| + | <p>The idea behind the tRNA amber supressor is to create an <em>artificial tRNA</em>, based on an existing tRNA that is loaded with a chosen amino-acid, and to change its anti-codon, replacing it by <em>the amber anti-codon</em>. By expressing this artificial tRNA in the cell, the ribosome will read-trhough the amber codon, and keep polymerasing the protein.</p> | ||
</td> | </td> | ||
| - | |||
<td> | <td> | ||
<img src="https://static.igem.org/mediawiki/2011/a/a3/Translation_short.gif"/> | <img src="https://static.igem.org/mediawiki/2011/a/a3/Translation_short.gif"/> | ||
| - | <center><p><u><b>Fig1:</b></u> | + | <center><p><u><b>Fig1:</b></u> Animation of the translation process (modified version of <a href="http://en.wikipedia.org/wiki/Ribosome">this animation</a>)</p></center> |
</td> | </td> | ||
</table> | </table> | ||
| - | <p>By creating | + | <p>By creating DNA that carries <zm>amber mutations</em> in the middle of its coding sequence, we create a protein that can be properly transcribed only if the artificial tRNA amber suppressor is present in the cell. Of course the mutations have to replace DNA coding for the same amino-acid that is on the artificial tRNA. This approach is sometime used in synthetic biology to create <em>artificial AND gates</em>.(Anderson 2007 <a href="https://2011.igem.org/Team:Paris_Bettencourt/tRNA_diffusion#references">[2]</a> and PKU Beijing team 2007 <a href="https://2011.igem.org/Team:Paris_Bettencourt/tRNA_diffusion#references">[3]</a>) </a> |
| - | < | + | <h2>An emitter/receiver design</h2> |
| - | |||
| + | <p><li>Here the <em>emitter cell</em> produces the mutated tRNA amber suppressor that will eventually travel through the nanotubes.</li> | ||
| + | <li>The <em>receiver cell</em> is then able to translate a protein (the T7 polymerase) containing the amber mutation. This protein triggers the reporter system.</li></p> | ||
| + | </html> | ||
| + | [[File:Trnaamber3.jpg|center|The tRNA diffusion system principle]] | ||
| + | <html> | ||
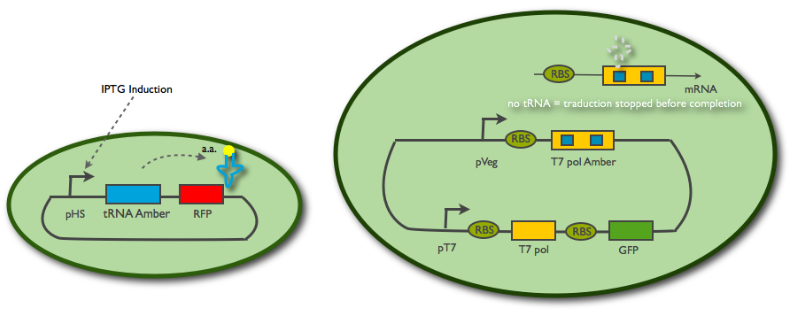
| + | <center><p><u><b>Fig1:</b></u> Scheme of the emitter cell construct (left) and the receiver cell contruct (right)</p> | ||
| + | </center> | ||
| + | <br /> | ||
| + | <h3>The emitter cell construct</h3> | ||
| - | < | + | <p>In order to measure the response time of the receiver cell, we needed to control the activity of the emitter cell. Therefore, we put the tRNA amber (BBa_K606034) under the control of an inducible promoter (pHyperSpank BBa_K606044). No RBS is necessary as it is a tRNA. A RFP was added downstream in order to monitor the expression of the tRNA.</p> |
| - | < | + | <h3>The receiver cell construct</h3> |
| - | < | + | <p>We want the receiver cell to be in a constantly "excitable" state, so we put a constitutive promoter(pVeg BBa_K143012) in front of the T7 amber(BBa_K606032). Indeed when the tRNA amber is not in the emitter cell, the ribosome will always stop the translation at the first codon stop (the amber mutation), so no T7 should be expressed. However, amber suppressing tRNAs entering the cell will allow the ribosome to finish the translation and a T7 RNA polymerase will be produced. The latter will then fix to a pT7 promoter(BBa_I719005) that we placed in front of a T7 gene(BBa_K145001) followed by a GFP gene(BBa_E1010). Therefore the first polymerase will induce an autoamplification system and an increase of green fluorescence.</p> |
| - | We | + | |
| - | |||
| - | |||
| + | <h2>Explanation step by step</h2> | ||
| + | |||
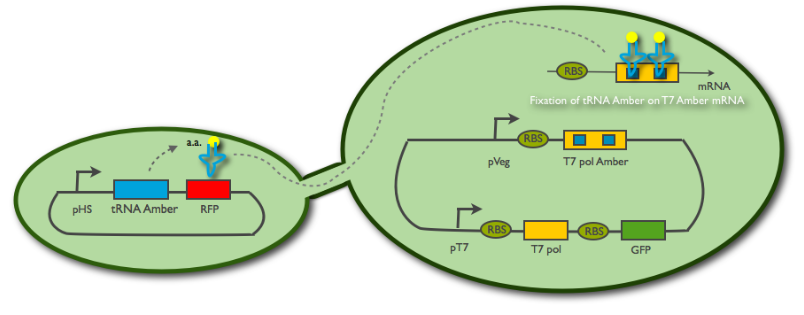
| + | <p><em>I.</em> The tRNA isn't expressed in the emitter cell, thanks to the Hyper Spank promoter. In the receiver cell, the translation of the T7 RNA polymerase is blocked by the amber codons in the sequence. It cannot be maturated and so the reporter system is silent. We launch the IPTG induction to produce amber tRNAs in the emitter cell, RFP fluorescence is also appearing.</p> | ||
| + | |||
| + | </html> | ||
| + | [[File:Trnaamber2.jpg|center|The tRNA diffusion system principle]] | ||
<html> | <html> | ||
| - | < | + | <center><p><u><b>Fig2:</b></u> When tRNA passes through the nanotube, it allows the receiver cell to translate the T7 polymerase</p> |
| + | </center> | ||
| + | <br /> | ||
| - | <p>The | + | <p><em>II.</em> The connection through the nanotube is established. Some tRNA diffuse into the receiver cell, and this is sufficient for the ribosome to read-through the two amber codons, thus polymerasing a few T7 RNA polymerase.</p> |
| + | <center> | ||
| + | <img src="https://static.igem.org/mediawiki/2011/e/ee/TRNA_amber_3.004-001.png" width=500px /> | ||
| + | <p><u><b>Fig3:</b></u> The T7 polymerase triggers the self amplifying reporter switch</p> | ||
| + | </center> | ||
| + | <br /> | ||
| + | <p><em>III.</em> Then, the T7 RNA polymerase is matured and becomes functional. It can trigger the pT7 promoter of the construct and the production of GFP.</p> | ||
| + | <center> | ||
| + | <img src="https://static.igem.org/mediawiki/2011/2/25/TRNA_amber_4.004-001.png" width=500px /> | ||
| + | <p><u><b>Fig4:</b></u> The positive feed back loop helps the signal to be amplified</p> | ||
| + | </center> | ||
| + | <br /> | ||
| + | |||
| + | <p><em>IV.</em> The T7 polymerase triggers the self amplifying switch by producing a few normal T7 RNA polymerases, which will keep self producing. In the mean time, because it is on the same mRNA, a proportional quantity of GFP will be produced.</p> | ||
| + | |||
| + | </html> | ||
| + | [[File:Trnaamber.jpg|center|The tRNA diffusion system principle]] | ||
<html> | <html> | ||
| + | <center><p><u><b>Fig5:</b></u> Complete scheme of the system</p> | ||
| + | </center> | ||
| + | <br /> | ||
| + | |||
| + | <p>The reporter system is active. We can look under the microscope. When a red cell (emitter cell) meets a dark cell (receiver cell) and that, a few minutes later, the receiver cell becomes green, we are sure that the message has passed through the nanotubes. | ||
| + | |||
| + | |||
| + | <h2>Models and experiments</h2> | ||
| + | |||
| + | <p>We have managed to build most of these constructs, and to model this system. We kindly invite you to visit the corresponding pages:</p> | ||
| + | <ul> | ||
| + | <li><em><a href="https://2011.igem.org/Team:Paris_Bettencourt/Modeling/tRNA_diffusion">Modeling</a></em></li> | ||
| + | <li><em><a href="https://2011.igem.org/Team:Paris_Bettencourt/Experiments/tRNA_diffusion">Results</a></em></li> | ||
| + | </ul> | ||
| + | <div id="citation_box"> | ||
| + | <p id="references">References</p> | ||
| + | <ol> | ||
| + | <li><i>tRNA determinants for transcription antitermination of the Bacillus subtilis tyrS gene</i>, F J Grundy, J A Collins, S M Rollins, and T M Henkin, available <a href="http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1369987/">here</a></li> | ||
| + | <li><i>Environmental signal integration by a modular AND gate</i>, J Christopher Anderson, Christopher A Voigt, and Adam P Arkin, available <a href="http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1964800/">here</a></li> | ||
| + | <li><i>Wiki of the 2009 PKU Beijing iGEM team, available </i> <a href="https://2009.igem.org/Team:PKU_Beijing/Project/AND_Gate_1/Design">here</a></li> | ||
| + | </ol> | ||
| + | </div> | ||
| + | <br> | ||
| + | |||
<!-- PAGE FOOTER -- ITEMS FROM COLUMN ! HAVE BEEN MOVED HERE -- RDR --> | <!-- PAGE FOOTER -- ITEMS FROM COLUMN ! HAVE BEEN MOVED HERE -- RDR --> | ||
| Line 101: | Line 154: | ||
</div> | </div> | ||
</body> | </body> | ||
| - | + | <div id="scroll_right"><a href="https://2011.igem.org/Team:Paris_Bettencourt/Modeling/tRNA_diffusion"><img src="https://static.igem.org/mediawiki/2011/e/e0/Arrow-right-big.png" style="width:100%;"></a><a href="https://2011.igem.org/Team:Paris_Bettencourt/Modeling/tRNA_diffusion">Model for tRNA</a></div> | |
| - | + | ||
| - | <div id="scroll_right"><a href="https://2011.igem.org/Team:Paris_Bettencourt/Modeling/tRNA_diffusion"><img src="https://static.igem.org/mediawiki/2011/e/e0/Arrow-right-big.png" style="width:100%;"></a><a href="https://2011.igem.org/Team:Paris_Bettencourt/Modeling/tRNA_diffusion"> | + | |
</html> | </html> | ||
Latest revision as of 10:01, 23 October 2011

The tRNA amber diffusion
The amber codon and the tRNA amber suppressor
|
The amber codon is one of the less used stop codon in bacteria. The principle of the artificial amber suppressor tRNA is to provide a tRNA corresponding to this stop codon. When the ribosome translates the RNA into a protein, it first looks for the RBS sequence and fixes on it. It tries to fit the codon it is located on with the three complementary bases of tRNA flying round. When it finds the correct tRNA with the anti-codon of the start codon with a methionine loaded on it, the translation starts. Then, codon after codon, the ribosome tries to fit many tRNA on the codon it is placed on, until it find the correct one, fixes the corresponding amino-acid and then moves to the next codon. When the ribosome reaches a codon that is sotp codon in the genetic code of the cell, the translation stops. The idea behind the tRNA amber supressor is to create an artificial tRNA, based on an existing tRNA that is loaded with a chosen amino-acid, and to change its anti-codon, replacing it by the amber anti-codon. By expressing this artificial tRNA in the cell, the ribosome will read-trhough the amber codon, and keep polymerasing the protein. |

Fig1: Animation of the translation process (modified version of this animation) |
By creating DNA that carries An emitter/receiver design
Fig1: Scheme of the emitter cell construct (left) and the receiver cell contruct (right)
The emitter cell construct
In order to measure the response time of the receiver cell, we needed to control the activity of the emitter cell. Therefore, we put the tRNA amber (BBa_K606034) under the control of an inducible promoter (pHyperSpank BBa_K606044). No RBS is necessary as it is a tRNA. A RFP was added downstream in order to monitor the expression of the tRNA.
The receiver cell construct
We want the receiver cell to be in a constantly "excitable" state, so we put a constitutive promoter(pVeg BBa_K143012) in front of the T7 amber(BBa_K606032). Indeed when the tRNA amber is not in the emitter cell, the ribosome will always stop the translation at the first codon stop (the amber mutation), so no T7 should be expressed. However, amber suppressing tRNAs entering the cell will allow the ribosome to finish the translation and a T7 RNA polymerase will be produced. The latter will then fix to a pT7 promoter(BBa_I719005) that we placed in front of a T7 gene(BBa_K145001) followed by a GFP gene(BBa_E1010). Therefore the first polymerase will induce an autoamplification system and an increase of green fluorescence.
Explanation step by step
I. The tRNA isn't expressed in the emitter cell, thanks to the Hyper Spank promoter. In the receiver cell, the translation of the T7 RNA polymerase is blocked by the amber codons in the sequence. It cannot be maturated and so the reporter system is silent. We launch the IPTG induction to produce amber tRNAs in the emitter cell, RFP fluorescence is also appearing.
Fig2: When tRNA passes through the nanotube, it allows the receiver cell to translate the T7 polymerase
II. The connection through the nanotube is established. Some tRNA diffuse into the receiver cell, and this is sufficient for the ribosome to read-through the two amber codons, thus polymerasing a few T7 RNA polymerase.

Fig3: The T7 polymerase triggers the self amplifying reporter switch
III. Then, the T7 RNA polymerase is matured and becomes functional. It can trigger the pT7 promoter of the construct and the production of GFP.

Fig4: The positive feed back loop helps the signal to be amplified
IV. The T7 polymerase triggers the self amplifying switch by producing a few normal T7 RNA polymerases, which will keep self producing. In the mean time, because it is on the same mRNA, a proportional quantity of GFP will be produced.
Fig5: Complete scheme of the system
The reporter system is active. We can look under the microscope. When a red cell (emitter cell) meets a dark cell (receiver cell) and that, a few minutes later, the receiver cell becomes green, we are sure that the message has passed through the nanotubes.
Models and experiments
We have managed to build most of these constructs, and to model this system. We kindly invite you to visit the corresponding pages:
References
- tRNA determinants for transcription antitermination of the Bacillus subtilis tyrS gene, F J Grundy, J A Collins, S M Rollins, and T M Henkin, available here
- Environmental signal integration by a modular AND gate, J Christopher Anderson, Christopher A Voigt, and Adam P Arkin, available here
- Wiki of the 2009 PKU Beijing iGEM team, available here
 "
"