Team:Harvard/Results
From 2011.igem.org
(→Bioinformatics) |
|||
| Line 5: | Line 5: | ||
__NOTOC__ | __NOTOC__ | ||
=Novel Zinc Fingers= | =Novel Zinc Fingers= | ||
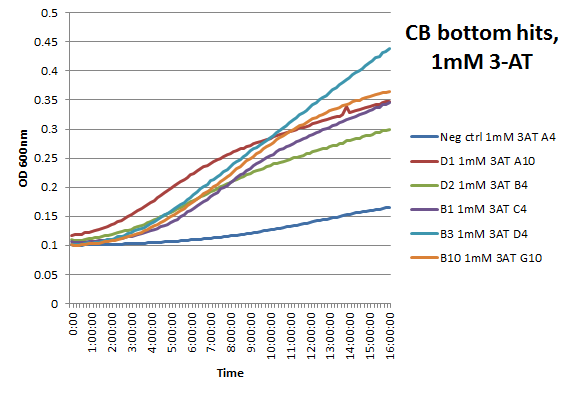
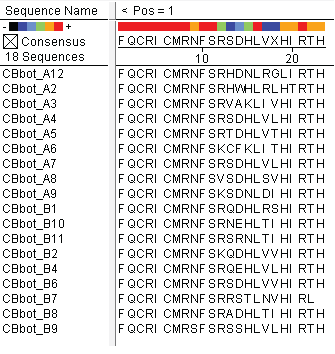
| - | + | We did not have time to test all 6 targets as planned: we chose to focus on zinc finger binders to DNA tripplet TGG, which is found in the color blindness gene bottom target. The TGG library was assembled and transformed into the selection strain with the proper binding site. So far we have screened 26 colonies and found 15 possible novel zinc finger binders to the sequence 5'-GTGGGATGG-3'. See [https://2011.igem.org/Team:Harvard/Results/Test#Color_Blindness_.28CB.29_Bottom_Hits:_15_Possible_Novel_Zinc_Fingers here] for more details. | |
| - | + | ||
| - | We did not have time to test all 6 targets as planned: we chose to focus on zinc finger binders to DNA tripplet TGG, which is found in the color blindness gene bottom target. | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
{| | {| | ||
|[[File: HARVcbbot_1mM.png|500px]] | [[File: HARVcb bot seqs.png|300px]] | |[[File: HARVcbbot_1mM.png|500px]] | [[File: HARVcb bot seqs.png|300px]] | ||
|} | |} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
=One-Hybrid Selection Strain= | =One-Hybrid Selection Strain= | ||
==Genome-Based Selection System== | ==Genome-Based Selection System== | ||
| - | We designed a one-hybrid metabolic system that was entirely genome-based. Using multiplex automated genome engineering (MAGE) and lambda red, we knocked out HisB, PyrF, and rpoZ; inserted a kanamycin cassette-zinc finger binding site-His3-URA3 construct into the 1529620 locus; and changed the zinc finger binding site directly on the genome. The strain was fully characterized and was sensitive enough to recognize a valid zinc finger when diluted as much as one into one million of negative controls. | + | We designed a one-hybrid metabolic system that was entirely genome-based. Using multiplex automated genome engineering (MAGE) and lambda red, we knocked out HisB, PyrF, and rpoZ; inserted a kanamycin cassette-zinc finger binding site-His3-URA3 construct into the 1529620 locus; and changed the zinc finger binding site directly on the genome. The strain was fully characterized and was sensitive enough to recognize a valid zinc finger when diluted as much as one into one million of negative controls. See [https://2011.igem.org/Team:Harvard/Results/Test#One-Hybrid_Selection_System here] for more details. |
[[File: HARVhybrid2_cropped.png|center|450px]] | [[File: HARVhybrid2_cropped.png|center|450px]] | ||
Revision as of 02:31, 18 October 2011
Overview | Biobricks | Source Code | Acknowledgments | Accomplishments
Novel Zinc Fingers
We did not have time to test all 6 targets as planned: we chose to focus on zinc finger binders to DNA tripplet TGG, which is found in the color blindness gene bottom target. The TGG library was assembled and transformed into the selection strain with the proper binding site. So far we have screened 26 colonies and found 15 possible novel zinc finger binders to the sequence 5'-GTGGGATGG-3'. See here for more details.
 | | 
|
One-Hybrid Selection Strain
Genome-Based Selection System
We designed a one-hybrid metabolic system that was entirely genome-based. Using multiplex automated genome engineering (MAGE) and lambda red, we knocked out HisB, PyrF, and rpoZ; inserted a kanamycin cassette-zinc finger binding site-His3-URA3 construct into the 1529620 locus; and changed the zinc finger binding site directly on the genome. The strain was fully characterized and was sensitive enough to recognize a valid zinc finger when diluted as much as one into one million of negative controls. See here for more details.
 "
"