Team:Arizona State/Notebook/August
From 2011.igem.org
(Difference between revisions)
Rubenacuna (Talk | contribs) m |
|||
| (5 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
__NOTOC__ | __NOTOC__ | ||
== Monday, August 1 == | == Monday, August 1 == | ||
| + | * Checked plates | ||
| + | :* Nothing | ||
| + | * Another RLT | ||
| + | :* This time with a vengeance | ||
| + | :* 2x Seq1, RA, DA in 1K3 | ||
| + | :* TSS cells | ||
| + | * Made more LB Kan broth | ||
| + | :* Because previous attempt was bad, ampR grew in it | ||
| + | * Tested TSS cells with puc19 | ||
| + | * Submitted BMES abstract | ||
== Tuesday, August 2 == | == Tuesday, August 2 == | ||
| + | * Transformation results from yesterday: | ||
| + | :* 4 colonies on seq1 | ||
| + | :* DA colonies all clumped together | ||
| + | :* RA fail, among stacks of failed transformations 3: | ||
| + | * New RLT for 2x | ||
| + | :* Varying DNA volumes added (1, 2.5, and 5 uL RLTs) | ||
| + | * PCR: | ||
| + | :* Seq1, RA, DA in puc57 using puc57 primers | ||
| + | ::* PCR settings: from previous | ||
| + | * Gel results: look the same as before | ||
| + | <br> [[Image:ASU_82_IMG_3989.jpg|400px]] | ||
| + | :* Clear bands ~100 for Seq1 | ||
| + | :* ~200 for DA, RA | ||
| + | * Submitted for sequencing: | ||
| + | :* BAHHH jason (sequencing delays) | ||
| + | :* Resubmitted 1x Seq1, RA, DA(pcr and prep), GFP, RFP, Cas3 | ||
| + | * Streak plates | ||
| + | :* BL21 DE3 (x2, one clearly better than the other) | ||
| + | :* 2xDA in pSB1K3 (clumped colonies initially) | ||
| + | * Liquid culture | ||
| + | :* 2xSeq1 in psb1k3 | ||
| + | :* BL21DE3 from glycerol stock | ||
| + | ::* Test tube with larger stopper may be contaminated | ||
== Wednesday, August 3 == | == Wednesday, August 3 == | ||
| + | * Liquid cultures | ||
| + | :* 2x Seq1 | ||
| + | :* 1xDA | ||
| + | * PCR | ||
| + | :* Cleanup of PCR amplified Seq1, RA, DA from puc57 | ||
| + | :* (We gelled this last night, looked good) | ||
| + | :* Used RA from this for RLT --> | ||
| + | * RLT | ||
| + | :* RA from PCR cleanup | ||
| + | * Prepared TSS competent cells | ||
| + | * Notes: | ||
| + | :* Decided not to continue with p. furiosus due to the cmr array's length (9kb) | ||
| + | :* Instead, we decided to explore our other options, like Type II streptococcus strains (which only have one cas protein involved in their mechanisms of silencing) | ||
== Thursday, August 4 == | == Thursday, August 4 == | ||
| + | * ABCDE PCR with the newest primers (v3) | ||
| + | :* 98deg initial denaturation for 30 seconds | ||
| + | :* Cycle {10 sec at 98 degrees | ||
| + | ::* Anneal at 63 degrees for 30 sec | ||
| + | ::* Elongate at 72 degrees for 130 sec} | ||
| + | :* 35 cycles | ||
| + | :* Then extension for 450 seconds | ||
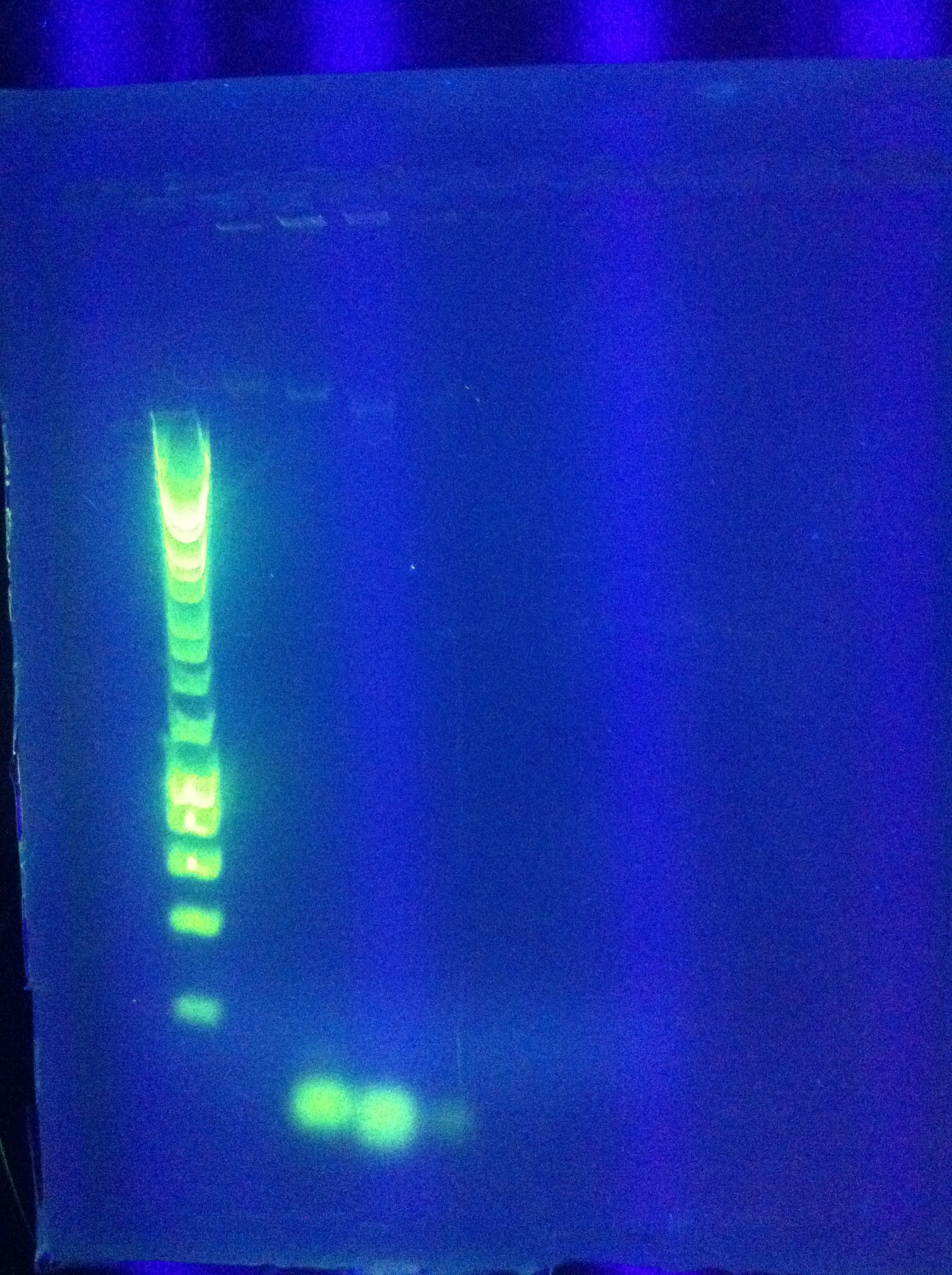
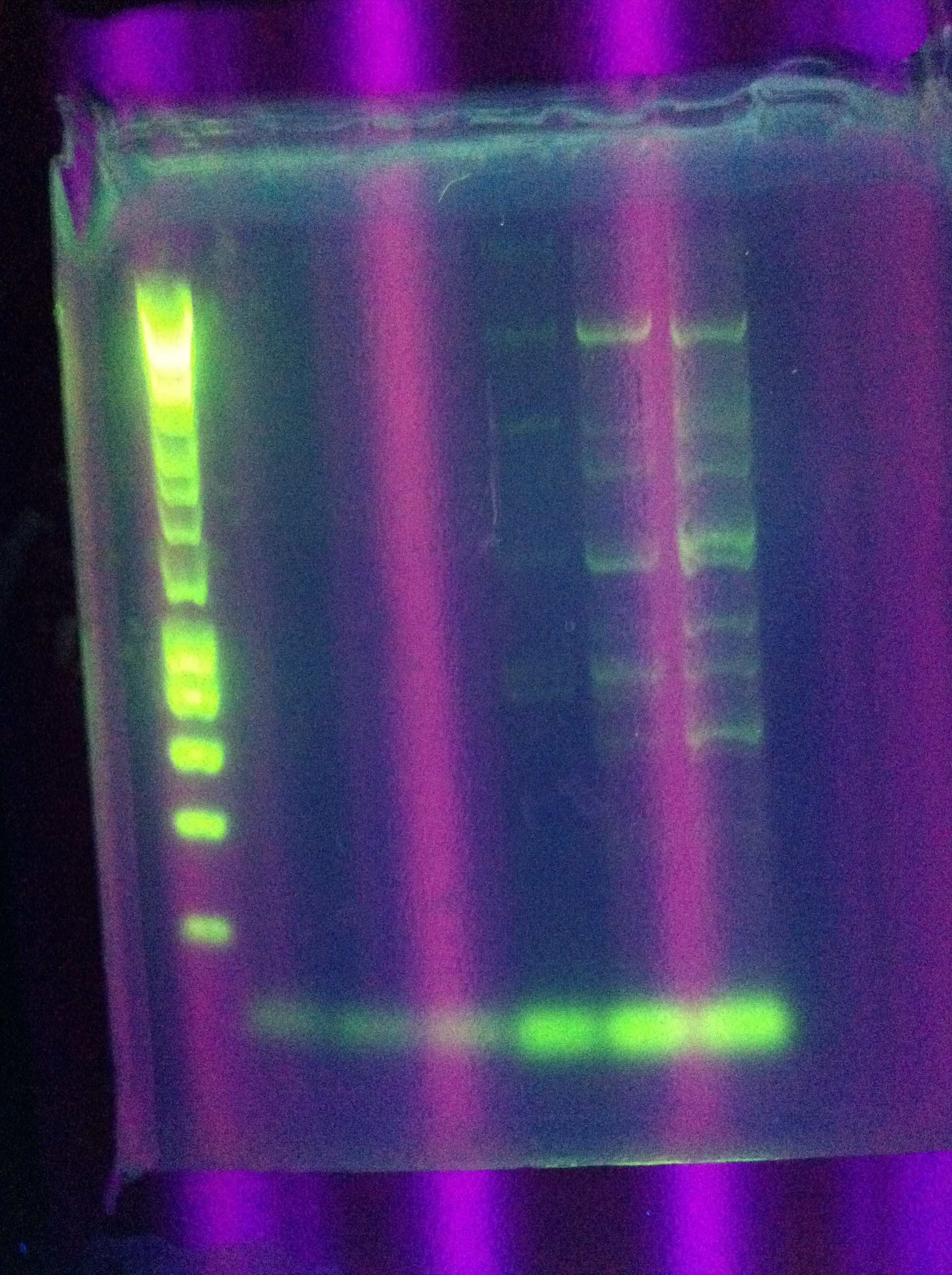
| + | * Results show clearly a band above where we would like it, at around 8kbp, and also some bands below the desired length at around 2kbp | ||
| + | <br> [[Image:ASU_84_Gel_for_PCR.jpg|400px]] | ||
| + | <br> [[Image:ASU_84_IMG_3998.jpg|400px]] | ||
== Friday, August 5 == | == Friday, August 5 == | ||
| - | + | * Miniprep: | |
| - | + | :* Using old protocol | |
| + | :* RAGE, SEGE, RFP overnight cultures | ||
| + | * Gel: | ||
| + | :* PCR from last night | ||
| + | ::* CasABCDE (upper): | ||
| + | :::* See photo | ||
| + | :::* Band > 10k, genomic? | ||
| + | ::* CasABCDE (lower): | ||
| + | :::* Same | ||
| + | * Gel extraction of PCR 8-4 (see photo) | ||
| + | * PCR of gel extraction CasABCDE: | ||
| + | :* Sample 1,2,3 | ||
| + | * Primers: CasABcDE_V3 F/R | ||
| + | :* Thermocycle: ABCDE802 | ||
| + | :* Template: gel extraction sample 1 of PCR 8-4 (above) | ||
| + | * Mg2+ gradient | ||
| + | :* Sample1: 0 mM | ||
| + | :* Sample2: 1mM | ||
| + | :* Sample3: 2mM | ||
| + | :* Cycle strarted 5:30pm | ||
| + | * Colony PCR: | ||
| + | :* Sample 1c, 2c,3c | ||
| + | :* Primers: CasABcDE_V3 F/R | ||
| + | * Thermocycle: ABCDE802 | ||
| + | :* Template: colonies from Jon's source plate | ||
| + | ::* 1c:BL21 DE3 -control | ||
| + | ::* 2c: BL21 DE3 -control | ||
| + | ::* 3c: Mg1655 +(hopefully we can amplify cas genes this way) | ||
| + | :* Cycle started 5:50pm | ||
| + | * New 10mM dNTP solution made from dNTP stock. | ||
| + | * RLT: | ||
| + | :* Todo | ||
== Monday, August 8 == | == Monday, August 8 == | ||
| + | GROUP MEETING | ||
| + | * Kylie's Update | ||
| + | :* Tried SLIC, not very successful | ||
| + | ::* (SLIC is a modification of Gibson) | ||
| + | :* New Ligation Method | ||
| + | ::* Idea is to cut with X, S | ||
| + | ::* This can then stick together multiple times, theoretically | ||
| + | ::* Different ligation times can allow for longer arrays (1x, 2x, 3x, 4x, 5x…) | ||
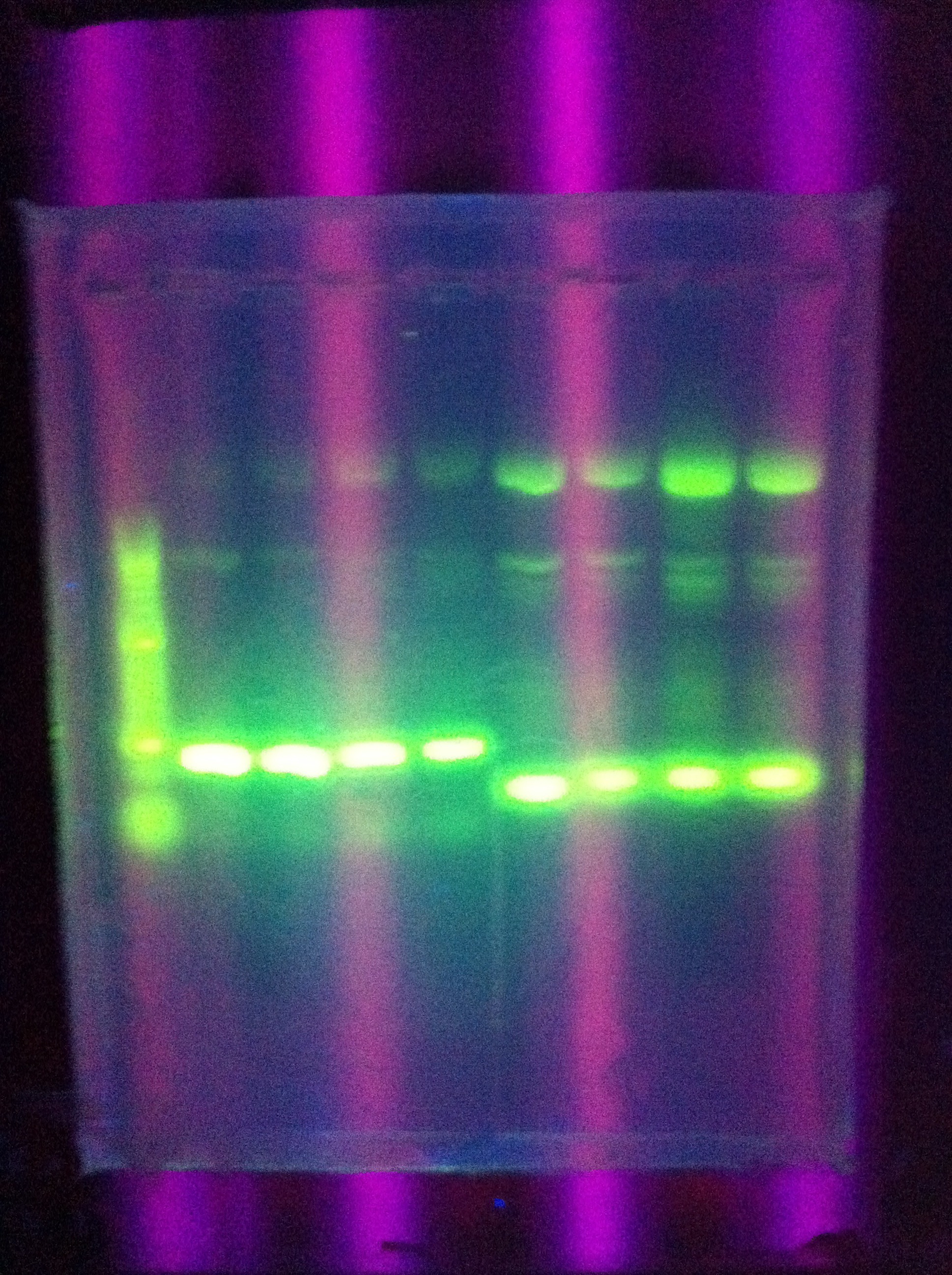
| + | ::* *Tried this for Seq1 | ||
| + | :::* Result: multiple bands at increasingly large lengths | ||
| + | <br> [[Image:ASU_88_IMG_4034.jpg|400px]] | ||
| + | <br> [[Image:ASU_88_IMG_4038.jpg|400px]] | ||
| + | ::::* w/normal ligation control to compare | ||
| + | ::* Consideration: could ligate "upside-down" - will this have any negative effect? | ||
| + | * Approach | ||
| + | :* RB + RA --> RSR or "Seq2" essentially | ||
| + | ::* Then, we can ligate using the KSB method | ||
| + | * Question: if there is an "upside-down" section, what even happens? | ||
| + | :* Is the complement side just skipped over? Will it hairpin? Will it mess something up? | ||
| + | :* Our best guess is that it will be fine, perhaps not every spacer will "work" but enough will | ||
| + | * What about self-targeting? | ||
| + | :* Sci tag prevents self targeting | ||
| + | * Organization | ||
| + | :* Personal Dropbox Lab Notebooks | ||
| + | * Specialization | ||
| + | :* Dan -- RB + RA, b. halodurans focus | ||
| + | :* Abhi -- "Seq2" assembly (XS method, etc) | ||
| + | :* Kylie -- Listeria innocua | ||
| + | :* Madeline -- CasABCDE, Cas3 (and put in Duet) | ||
| + | :* Keith -- Ginkgo | ||
| + | :* Nisarg -- Ginkgo | ||
| + | :* Ryan -- CasABCDE, Cas3 | ||
| + | :* Ruben -- drylab | ||
| + | :* Ethan -- drylab | ||
| + | :* Joseph -- XS method (seq1, RA) | ||
| + | * To order | ||
| + | :* Gloves | ||
| + | :* Plates | ||
| + | :* T4 polymerase | ||
| + | :* Miniprep | ||
| + | :* Listeria innocua items | ||
== Tuesday, August 9 == | == Tuesday, August 9 == | ||
| + | * Compiled cas gene information to table on wiki | ||
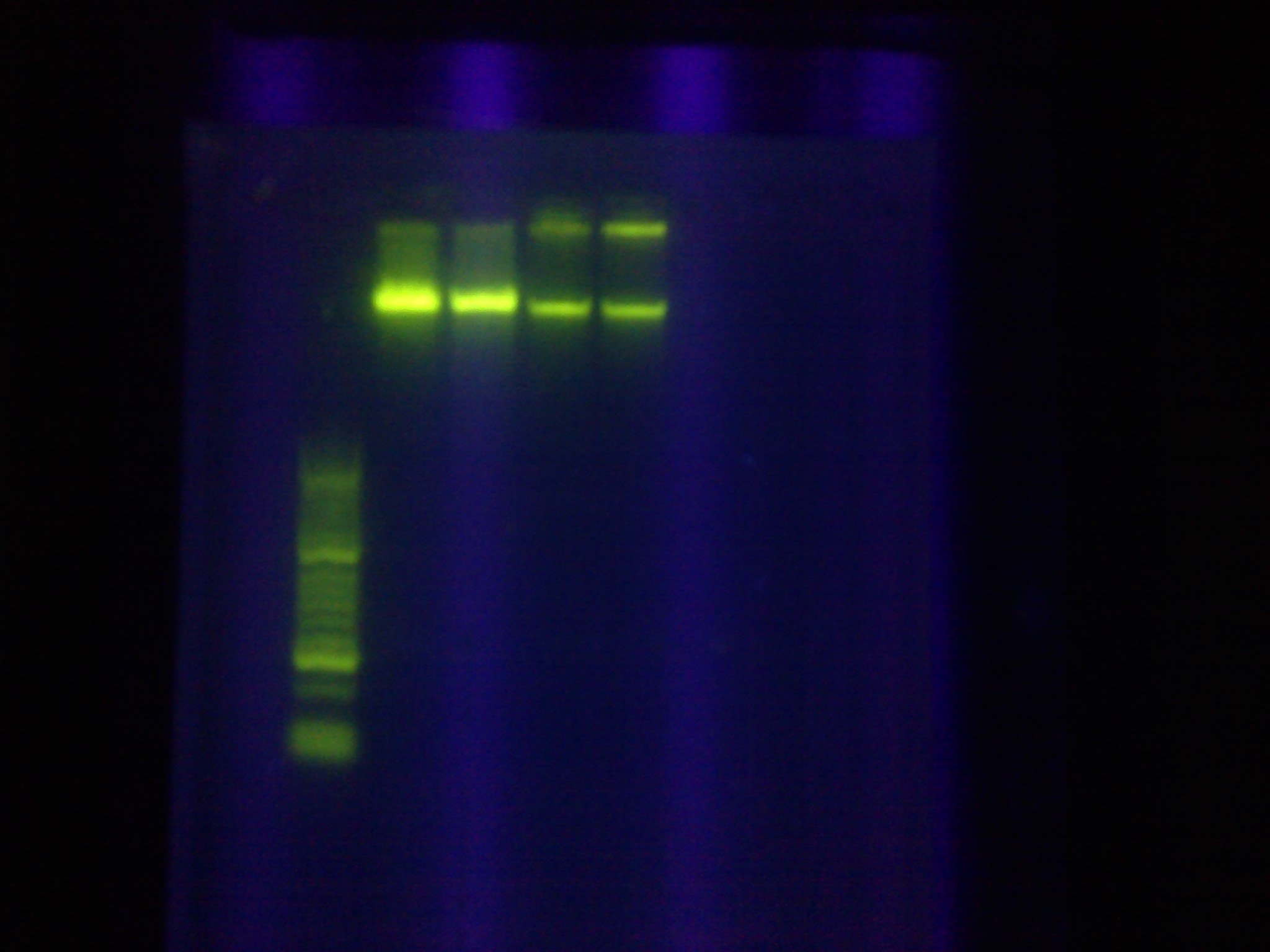
| + | * Confirmed that RA was successfully amplified with a gel | ||
| + | <br> [[Image:ASU_89_IMG_4047.jpg|400px]] | ||
| + | * Proceeded to prepare XS restriction/ligation for RA | ||
| + | * Actual reagent volumes for 50 uL restriction/ligation can be found in lab notebook under 8-9-11 | ||
| + | * Incubated restriction/ligation at 37 degrees overnight | ||
| + | * Liquid cultures from 8/8 failed to grow | ||
| + | :* Potentially bad LB-Kan broth | ||
| + | * Made new LB-Kan broth | ||
| + | * New liquid cultures of Seq1 and RA (2x, each) | ||
| + | :* Old LB-Kan broth | ||
| + | :* New LB-Kan broth | ||
| + | TO DO for 8-10-11: | ||
| + | * Gel RL results | ||
| + | * PCR cleanup restriction/ligation results | ||
| + | * Possibly PCR cleanup RA inserts from PCR amplification for re-restriction/re-ligation | ||
== Wednesday, August 10 == | == Wednesday, August 10 == | ||
| + | * Liquid cultures from 8/9 all failed. | ||
| + | :* Both old and new LB+Kan broth | ||
| + | * Positive and negative controls started | ||
| + | :* Old and new LB-Kan broth | ||
| + | * RA and Seq1 from 8/8 streak plated | ||
| + | * Liquid cultures in LB-Amp broth started | ||
| + | :* From pUC57, RA and Seq1 | ||
| + | :* 2 liquid cultures of each | ||
| + | * Gel results for last night's PCR of Cas3 (see pic) | ||
| + | :* ((recall settings: Cas3724 protocol, which is the previously successful run)) | ||
| + | :* Very bright nonspecific band around 1000 | ||
| + | :* Lots of small bright bands at low lengths (200-600) | ||
| + | :* Band at the top near 10k | ||
| + | :* MAYBE a slight hint of a band around where we want it, but not much there | ||
| + | * Discussion: | ||
| + | :* I am not sure why we are not seeing the same result that we saw on 7/24. | ||
| + | :* Perhaps trying a higher elongation time would help. | ||
| + | :* I will compile a document with our PCR history, perhaps this will help us figure things out. | ||
| + | * Really, we need to be doing this over and over in a day. Multiple runs instead of just one. Gotta get to that point. | ||
| + | * Cas3 attempt | ||
| + | :* Same settings, used 26ng/uL template instead as this is the template that was used back on 7/24 [Sample ID: PRANCER] | ||
| + | * Gel results (see pic) | ||
| + | :* Seeing a nonspecific-ish area around 1000, also lots of smaller bands | ||
| + | * Ran another PCR for ABCDE, same settings as before but one degree higher for annealing temp. Also added DMSO. [sample ID: PRANQSTER] | ||
| + | * Ligated sample 4L (Seq1) to pSB1A3 linearized backbone cut with XbaI and SpeI then deposphorylated. | ||
| + | * Samples 4L 1A3 1 and 4L 1A3 2 (12ul of 4L and 2ul of 1A3 in 20ul reaction) | ||
| + | * Negative control 1A3 L | ||
| + | * Ligase heat inactivated by "OVERKILL" protocol for 10 minutes. | ||
| + | * Samples stored at -20 degrees on restriction ligation section of tray. | ||
== Thursday, August 11 == | == Thursday, August 11 == | ||
| + | * Ran a gel of last night's PCR of ABCDE (see pic) | ||
| + | :* DMSO worked -- same results as previously but less overall bands | ||
| + | :* Seemed to hone in on the band that is above our desired length (~8k or so) | ||
| + | * (For reference to the picture, we ran some kind of restriction product that I can't recall on the right side w/ Hyperladder II) | ||
| + | :* Does anyone remember this? Might be Restricted RB | ||
| + | * Liquid cultures of leader sequences | ||
| + | :* One colony from each of the 4 plates | ||
| + | ::* Leader Seq MG1655 Dilute | ||
| + | ::* Leader Seq MG1655 Concentrated | ||
| + | ::* Leader Seq B. halo Dilute | ||
| + | ::* Leader Seq B. halo Concentrated | ||
| + | * Temperature Gradient PCR (Madeline and Ryan) | ||
| + | :* CasABCDE | ||
| + | ::* MgCl2 --> 2uL, 4uL, 5uL | ||
| + | ::* Temp --> 60, 63, 66, 69 | ||
| + | :* Cas3 | ||
| + | ::* MgCl2 --> 2uL, 4uL, 5uL | ||
| + | ::* Temp --> 60, 62, 64, 66 | ||
| + | * Gel Result (see pic) | ||
| + | :* Ran MgCl210 for each of the 8 temp rows (4ea for ABCDE, Cas3) | ||
| + | :* No bands for ABCDE 3: | ||
| + | :* Similar results that we have been getting for Cas3 | ||
| + | ::* 60 and 62 have bands less than our desired target, up to perhaps 1500 | ||
| + | ::* 64 had a couple of bands above this, perhaps one faint one near our 2600 target | ||
| + | ::* 66 had more nonspecific amplification, and no clear bands in our target range | ||
| + | * Moving forward | ||
| + | :* Nest for ABCDE, we will see how this goes | ||
| + | :* Cas3 we'll have to keep trying…perhaps it may be worth it to try DMSO, longer elongation time, something. Maybe Taq. | ||
== Friday, August 12 == | == Friday, August 12 == | ||
| + | * Visit to Bioscience HS | ||
| + | :* Madeline, Abhi, Nisarg | ||
| + | :* Took a little tour of the school, saw their lab space (which is pretty divine) | ||
| + | ::* Fun tidbit: each room has a plaque outside with the room number and an element name for the type of room (ex: O for office, He for men's restroom, Fe for women's restroom, etc..) | ||
| + | :* Spoke w/ Ms. Singh's biotech class + some interested sophomores and seniors | ||
| + | :* They will likely start an iGEM team, they seem quite interested | ||
| + | ::* Juan will be captain | ||
| + | :* Singh will contact us soon after they speak with the administration about how it will all fit in and what our role will be | ||
| + | * PCR using Nest primers for ABCDE | ||
| + | :* Tm1: 74, Tm2: 77 --> Anneal at 72 deg (higher than this is not recommended) | ||
| + | :* Used 2-step Phusion protocol | ||
| + | ::* 98 deg for 30 sec | ||
| + | ::* {98deg for 10 sec, 72 deg for 2:30} X 35 cycles | ||
| + | ::* Extension for 7:30 at 72 deg | ||
| + | ::* Held at 4deg until run was cancelled | ||
| + | * Gel results (see pic) | ||
| + | :* The 8x MgCl2 tube has the best result | ||
| + | :* Nice clear bands at around 600, 1k, 2k, 4k, 6k, 8k | ||
| + | :* PLAN: use this as template for ABCDE PCR | ||
| + | ::* Use 1uL per tube (note, we don't know the concentration of the strand we want as template, so this is our best guess as to what would get us between 1pg and 10ng as recommended by the Phusion protocol for non-genomic DNA) | ||
| + | * Also today: | ||
| + | :* PCR amplification of Seq1, RA | ||
| + | ::* 14 tubes total (7 each at null, 0, 1, 2, 4, 8, 10) | ||
| + | :* Note we are running out of Phusion already! | ||
| + | * Another PCR (Madeline and Kylie) | ||
| + | :* Using template from earlier PCR | ||
| + | :* 3 tubes, all had 8x MgCl2 (4 uL) | ||
| + | :* Titration of template concentration: 1/5, 1/50, 1/500 | ||
| + | ::* Note, for 1/500 we ran out of Phusion (saddest bear 3=) so we used Taq | ||
| + | :* Protocol | ||
| + | ::* Same as ABCDE808 but 35 cycles total (2:10 elongation time, 63deg annealing temp) | ||
== Saturday, August 13 == | == Saturday, August 13 == | ||
| + | * Restriction on poly x to remove incorrectly oriented pieces | ||
| + | * Gel results for Taq Polymerase try at Cas3 (see picture) | ||
| + | :* Old primers: nonspecific all the way down | ||
| + | :* Newer primers (v2): a series of bands all below 1000bp | ||
| + | * Other things in lab: | ||
| + | :* XS Restriction after overnight ligation | ||
| + | :* 8/8 plate growth for Keith's Ginkgos | ||
| + | :* Liquid cultures of 2x attempts, Dan's RABs | ||
| + | :* Miniprep of leader sequences & nanodrops (maybe this was Friday) | ||
== Sunday, August 14 == | == Sunday, August 14 == | ||
| + | * Genome prep MG1655 | ||
== Monday, August 15 == | == Monday, August 15 == | ||
| + | Team meeting | ||
| + | * Updates | ||
| + | :* Cas genes - not looking good, ran out of Phusion, will keep trying w/ some adjustments | ||
| + | :* Ginkgo woes | ||
| + | * Today | ||
| + | :* Liquid culture of MG1655 | ||
| + | ::* to make it competent | ||
| + | :* Sequencing | ||
| + | ::* Leader Sequences | ||
| + | ::* 2x RA | ||
| + | ::* Duet | ||
| + | ::* Whatever else is on the board | ||
| + | :* Streak plate pRSF Duet (old mini prep, could be weird) | ||
| + | :* New media, new plates | ||
| + | :* Streak plate polySeq1 | ||
| + | ::* Liquid culture later, mini prep tonight or tomorrow | ||
| + | :* Cas3 PCR w/ uber long elongation | ||
| + | ::* Look again at annealing temps | ||
== Tuesday, August 16 == | == Tuesday, August 16 == | ||
| + | * Liquid cultures | ||
| + | :* Kylie's 4L1A3 (orig. XS ligation of Seq1 in pSB1A3 | ||
| + | :* 1655 from glycerol stock for genome prep and competency | ||
| + | :* BL21 DE3 for more TSS cells | ||
| + | :* Keith's 2x RA and Seq1 streak plates from 8/15 (grew this time!) | ||
| + | ::* Used LB-Kan broth from 8/11 (made by Abhi) | ||
| + | * PCR amplification of Seq1 in pUC57 | ||
| + | :* Gel visualized, ~150bp bands in samples 4,8,10 | ||
| + | ::* Still on transilluminator | ||
| + | :* Samples labeled and in -20 | ||
| + | * PCR log updated | ||
| + | * New PCR of Cas genes | ||
| + | :* Going to try Cas3 with 7/24 settings with OLD PRIMERS | ||
| + | :* Also, try an ABCDE Nest with 8x MgCl2 (this was good before) and a small temperature gradient | ||
| + | ::* We can then use the result as a template (and perhaps sequence it for funzies to see if we're getting it) | ||
| + | :* ((more details in Madeline's folder, including results, though pic is here too)) | ||
| + | * PCR | ||
| + | :* Cas3, 7/24 protocol, OLD PRIMERS | ||
| + | ::* (we have been using the more recent primers…) | ||
| + | ::* MgCl2 gradient | ||
| + | :* ABCDE Nest, 8/13 protocol but with temperature gradient from 70 to 74 | ||
| + | * Results: see picture | ||
| + | :* Comparable results for Cas3 to the original run…will gel this by itself tomorrow | ||
| + | :* Nest does not look very good, especially compared to 8/13 result | ||
| + | ::* Might just use 8/13 tube as template, try again | ||
== Wednesday, August 17 == | == Wednesday, August 17 == | ||
| + | * Made tss cells (bl21, mg1655) | ||
| + | * Genome prep: mg1655 | ||
| + | * 2 Minipreps of the K12 Leader and the B. Halo Leader. | ||
== Thursday, August 18 == | == Thursday, August 18 == | ||
| + | * Gel of the Cas3 and Nest ABCDE | ||
| + | * Sent RARB and RA 2X for sequencing | ||
== Friday, August 19 == | == Friday, August 19 == | ||
| + | *Synopsis: | ||
| + | :* Seq1 PCR's 5 samples are for both SLIC, Poly-X ligation and 1X samples. | ||
| + | :* New LB-Amp made. plus a negative control. | ||
| + | :* K12 Leader/Seq1 ligations incubated and will be ready for transformation on Saturday 8-20. | ||
| + | :* Microcentrifuge tubes, PCR tubes, and Test tubes autoclaved, since nuclease contamination has been present among samples. | ||
| + | :* Sterile p10 tips autoclaved and ready to use. | ||
| + | * K12 Leader Sequence dephosphorylated | ||
| + | * Ligations of K12 Leader and 1x Seq1 | ||
| + | *Samples: | ||
| + | :* L1: K12+Seq1 (no PCR cleanup for Seq1) | ||
| + | :* L2: K12+Seq (W/PCR cleanup for seq1 using Quiagen PCR cleanup kit) | ||
| + | :* L3: K12+Seq (W/PCR cleanup for seq1 using Quiagen PCR cleanup kit) | ||
| + | :* L4: Negative control, phosphatased vector only | ||
| + | * Incubated at 16 degrees for 12 hours. | ||
| + | * Media Prepared, since negative controls of LB-AMP Dan/Abhi 8-15 grew with widltype K12 MG1655. | ||
| + | :* 400mL of LB broth prepared and boiled in microwave using "Frozen Entree" plus a couple minutes, sample was returned to room temperature then 400ul of 1000x Ampicillin added. | ||
| + | :* Negative control ran with widltype K12 MG1655. | ||
| + | * PCR of Seq1: | ||
| + | :* 5 reactions of Seq1 PCR off pUC57 prepared and ran. Samples are for both SLIC, Poly-X ligation and 1X samples. | ||
== Saturday, August 20 == | == Saturday, August 20 == | ||
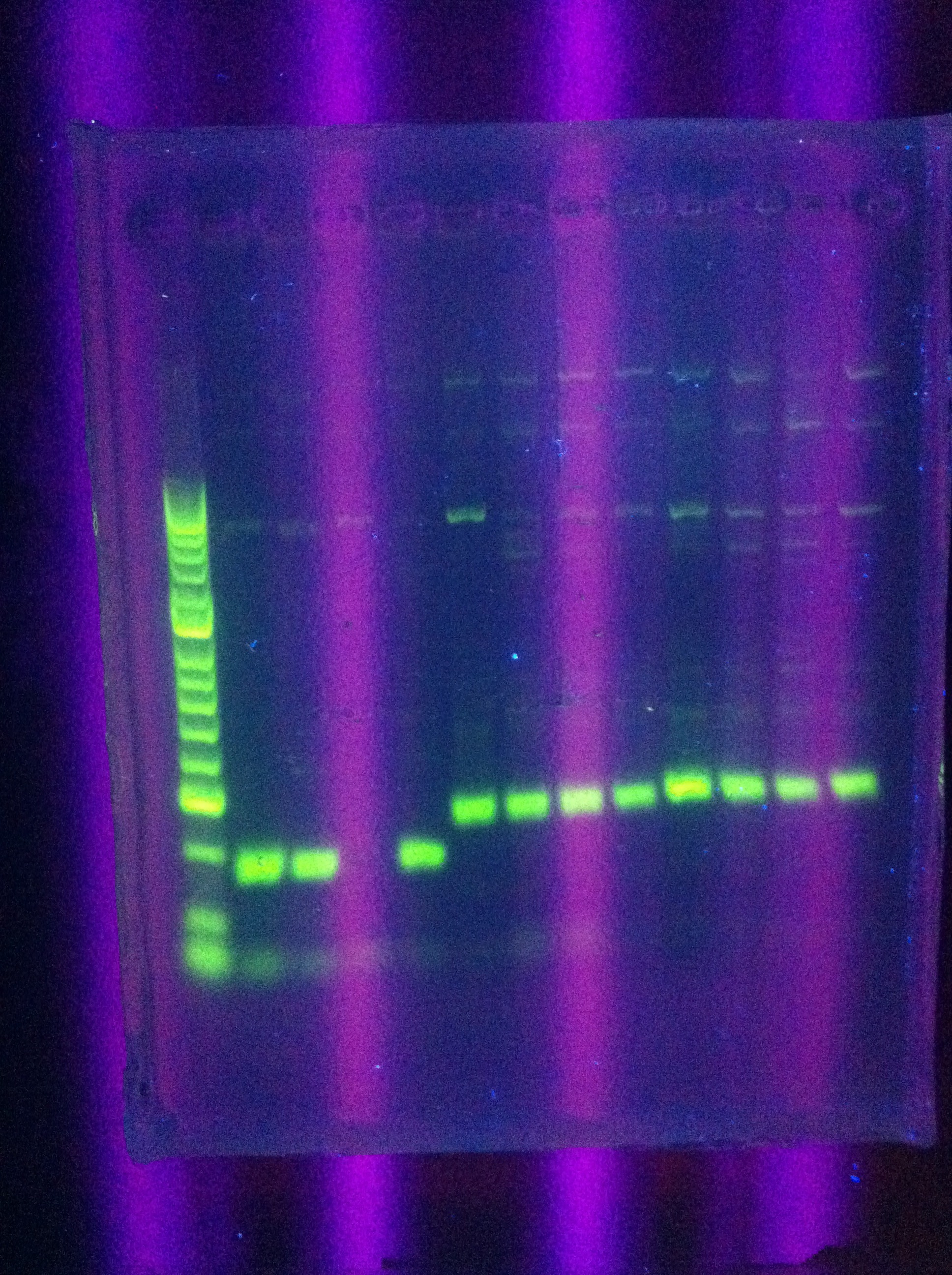
| - | + | * Restriction of GFP in 1A3 and RFP in 1K3 | |
| - | + | Incubated for 2 hours at 37 degrees. | |
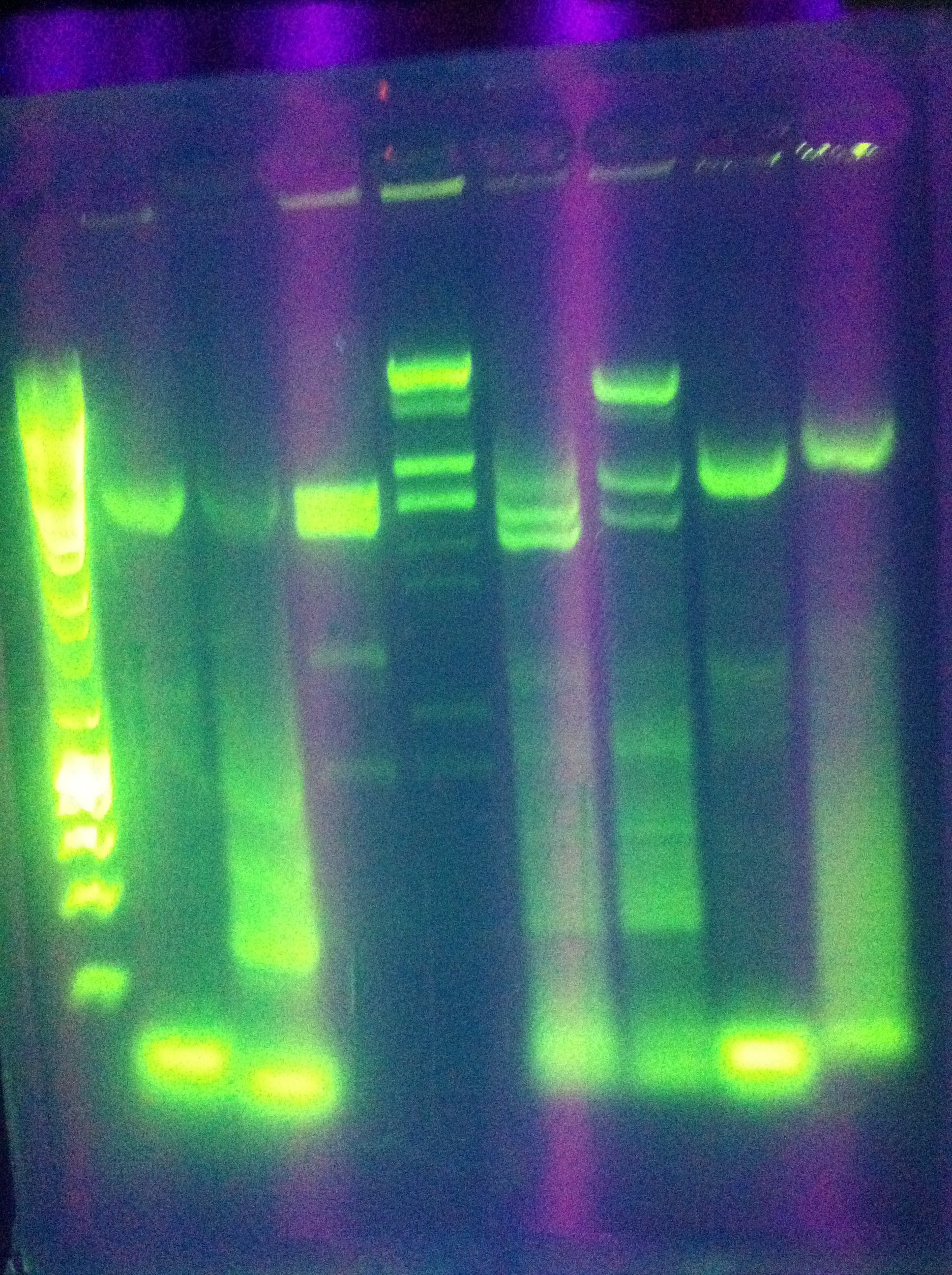
| - | + | * Diagnostic gel ran of GFP and RFP cut. GFP shows slight degradation, with less linearized plasmid than expected. RFP backbone not visible. Seq1-1 PCR product from 8-19 is present at ~200bp. GFP in 1A3 and RFP in 1K3 on the DAN/Abhi tray do not pass. | |
| + | :* Lane 1: Hyperladder 1 | ||
| + | :* Lane 2: GFP in pSB1A3 | ||
| + | :* Lane 3: GFP cut with EcorI and PstI | ||
| + | :* Lane 4: RFP in pSB1K3 | ||
| + | :* Lane 5: RFP cut with EcorI and PstI | ||
| + | :* Lane 6: K12 Leader 2 (from 8-18) | ||
| + | :* Lane 7: K12 Leader 4 (From 8-18) | ||
| + | :* Lane 8: Seq1 PCR 1(From 8-19) | ||
| + | * Restrictions of RA and Seq1 | ||
| + | :* Samples: (Stored at -20 on top shelf "1-5 KSB") | ||
| + | ::* 1: Seq1 1 (From 8-19) | ||
| + | ::* 2: Seq1 2 (From 8-19) | ||
| + | ::* 3: RA PCR (Dan provided sample) | ||
| + | ::* 4: RA 0.1 (Dan provided sample) | ||
| + | ::* 5: RA 8,10 (Dan provided sample) | ||
| + | * Note: Dan's samples where restricted given his protocol overnight at 37 degrees, then samples where accidentally re-restricted by myself; samples where promptly heat inactivated following addition of restriction enzyme. | ||
| + | * Samples 1 through 5 were then cleaned with the Quiagen PCR Cleanup kit spin columns and eluted with 50uL Nuclease free H2O, instead of Elution Buffer (EB) provided by protocol, since EB is known to interfere with downstream enzyme reactions. | ||
| + | * Coupled Restriction Ligation Attempt | ||
| + | :* Sample incubated for 1 hour at 25 degrees then 8uL was ran on diagnostic gel. Sample shows nebulization at ~400 bp, this may be nuclease contamination but may not be. It may be a mixture of products forming from multiple ligations. Or it may be an unknown nuance inhibiting mutual restriction and ligation (See image: 8-20 1_2_3_4_5_RE+Lig). Note: the sample may need to be restricted, then column prepared using the Sigma-Aldrich PCR Cleanup Kit (Which appears to more effectively remove small <100nt fragments than Quiagen kit), then attempt coupled restriction ligation. If nebulazation still appears the reaction concept may be flawed. | ||
| + | |||
== Wednesday, August 24 == | == Wednesday, August 24 == | ||
| + | * Minipreps: | ||
| + | :* RARB A1 Kan (1 total) | ||
| + | :* GFP Gen 1, GFP Gen 2, RARB LB Kan (3 total) | ||
| + | :* All 4 sent to nanodrop | ||
| + | * Gels | ||
| + | :* Gel 1: pIDT + Seq1 + Leader (E) [4 lanes], pIDT + Leader [2 Lanes] | ||
| + | ::* Both samples in the gel look good. | ||
| + | :* Gel 2: CasABCDE [3 lanes], Cas3 [4 lanes] | ||
== Thursday, August 25 == | == Thursday, August 25 == | ||
| + | * Listeria received from ATCC. | ||
| + | :* Glass vial was opened and bacterial pellet was eluted with 500uL of Brain-Heart Infusion composition broth. The pellet was given 1 minute to dissolve, the 500uL was then placed in 5mL of Brain-Heart Infusion composition broth (Sample: KSB BHI 8-25 Listeria, Media: BHI KSB 8-24). 200uL was transfered to a BHI agar plate and both the 5mL tube and BHI plate where incubated at 37degrees overnight. | ||
| + | * One streak plate of BL21(DE3) and K12 MG1655 where prepared and incubated at 37 degrees overnight. | ||
| + | * Two liquid cultures where prepared from Jon's source plate, one of BL21(DE3) and the other of K12 MG1655. Samples were incubated over night. | ||
| + | * Tomorrow: | ||
| + | :* TSS cells should be prepared. | ||
| + | :* BL21 Should be made into glycerol stocks and TSS cells. | ||
| + | :* MG1655 Should be made into glycerol stocks and genomic prepped. | ||
| + | :* Listeria should be re-plated, transfered to new media, and made into glycerol stocks. | ||
== Friday, August 26 == | == Friday, August 26 == | ||
| + | * CULTURES: | ||
| + | :* Listeria re-streaked, glycerol stocks prepared in both 20% glycerol and 50% glycerol concentrations (Labelled on vials). | ||
| + | ::* Samples 1-12 20% Glycerol | ||
| + | ::* Samples 13-16 50% Glycerol | ||
| + | * GENOMIC DNA: | ||
| + | :* Genomes of both K12 MG1655 and Listeria innocua Prepared using Sigma-Aldrich genome preparation kit (protocol for gram-negative). | ||
| + | ::* Samples MG1655 1 and LI 1 where eluted in nuclease free water. | ||
| + | ::* Sample MG1655 2 and LI 2 where eluted in elution buffer. | ||
| + | * Sample nanodrop results: | ||
| + | :* MG1655 1 (H2O): 38.4 ng/ul | ||
| + | :* MG1655 2 (Elution Solution): 26.8 ng/ul | ||
| + | :* LI 1 (H2O): 77.5 ng/ul | ||
| + | :* LI 2 (Elution Solution): 51.3 ng/ul | ||
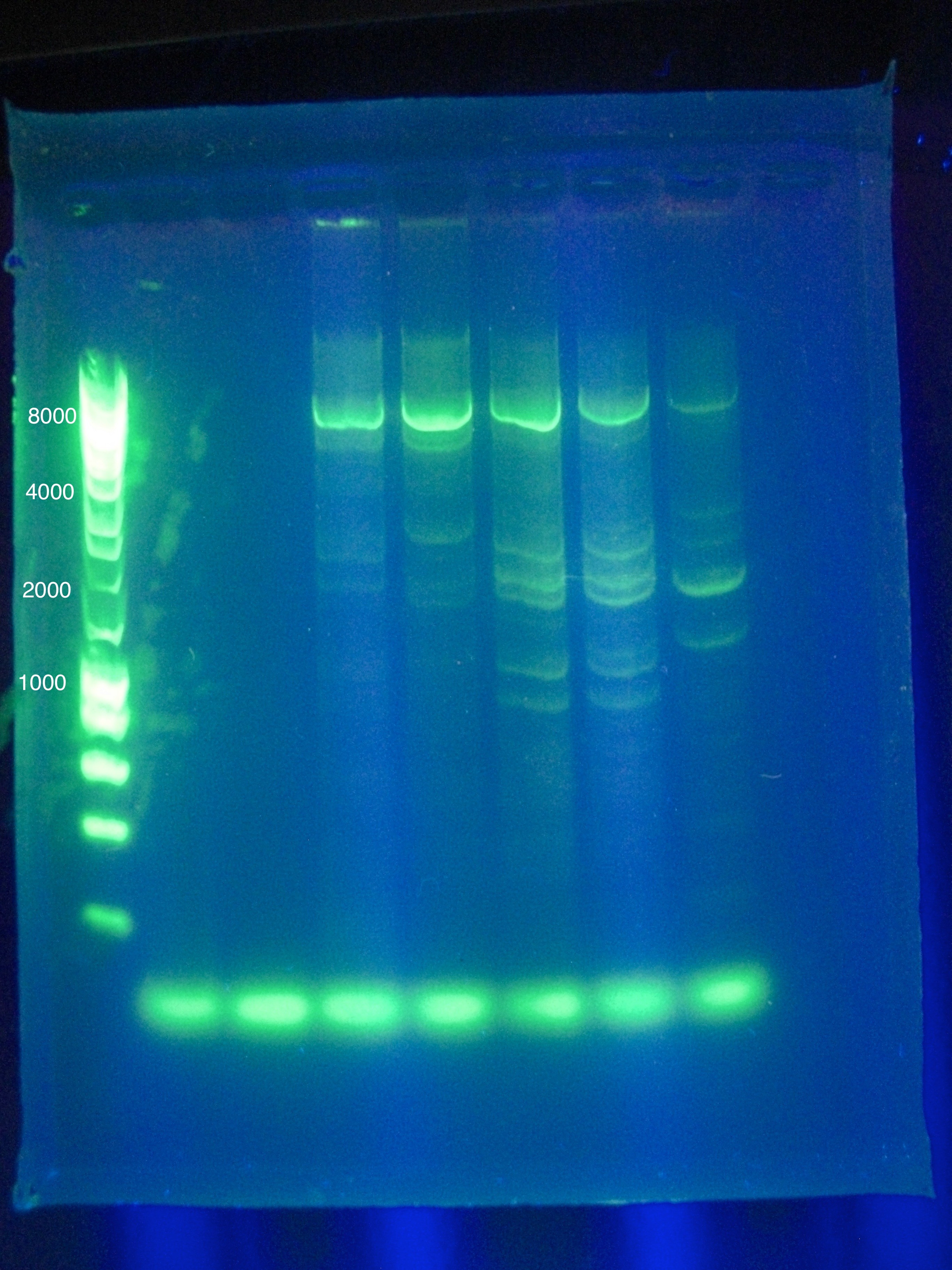
| + | * Whole Genomes were visualized on a 1% agarose gel, with the goal of visualizing if there is genome fracturing or degradation. | ||
| + | :* Lane 1: Hyperladder1 | ||
| + | :* Lane 2: MG1655 1 (H2O) | ||
| + | :* Lane 3: MG1655 2 (Elution Solution) | ||
| + | :* Lane 4: LI 1 (H2O) | ||
| + | :* Lane 5: LI 2 (Elution Solution) | ||
| + | <br> [[Image:ASU_26_Genome_Listeria.jpg|400px]] | ||
| + | * PRIMERS: | ||
| + | :* 16s rRNA Forwar/Revers primers recieved and diluted to 100uM concentrartion | ||
| + | :* M13 Forward (-20) and M13 Reverse (-27) primers recieved and diluted to 100uM concentration. | ||
| + | * PCR: | ||
| + | :* The aforementioned genome preparations where ran with a positive control PCR using the 16s rRNA primers. | ||
| + | :* Using a magnesium gradient set up by Ethan. | ||
| + | * Results visualized by 1% agarose gel (8-27) | ||
== Saturday, August 27 == | == Saturday, August 27 == | ||
| + | * More Genomic DNA: | ||
| + | :* Using sigma-aldrich genomic DNA preparation kit | ||
| + | * NEW -80 DEGREE BOX: | ||
| + | :* "Kylie Glycerol Stocks" | ||
| + | ::* Currently contains (Source): listeria stocks 8-25/8-26/8-27 (ATCC), BL21DE3 1-16(Misra), DH10B (NEB) | ||
| + | * Transformation of LI_T7LRSR (KanR) and LI_RSR (AmpR) using NEB competent sells and NEB protocol. | ||
== Sunday, August 28 == | == Sunday, August 28 == | ||
| - | + | * New Listeria Innocua CLIP11262 genome preparations using Sigma-Aldrich Genomic DNA Preparation kit. | |
| - | + | :* Samples 1-6, 2-28 | |
| + | * Note: All eluted in kit Elution Buffer. | ||
| + | |||
== Wednesday, August 31 == | == Wednesday, August 31 == | ||
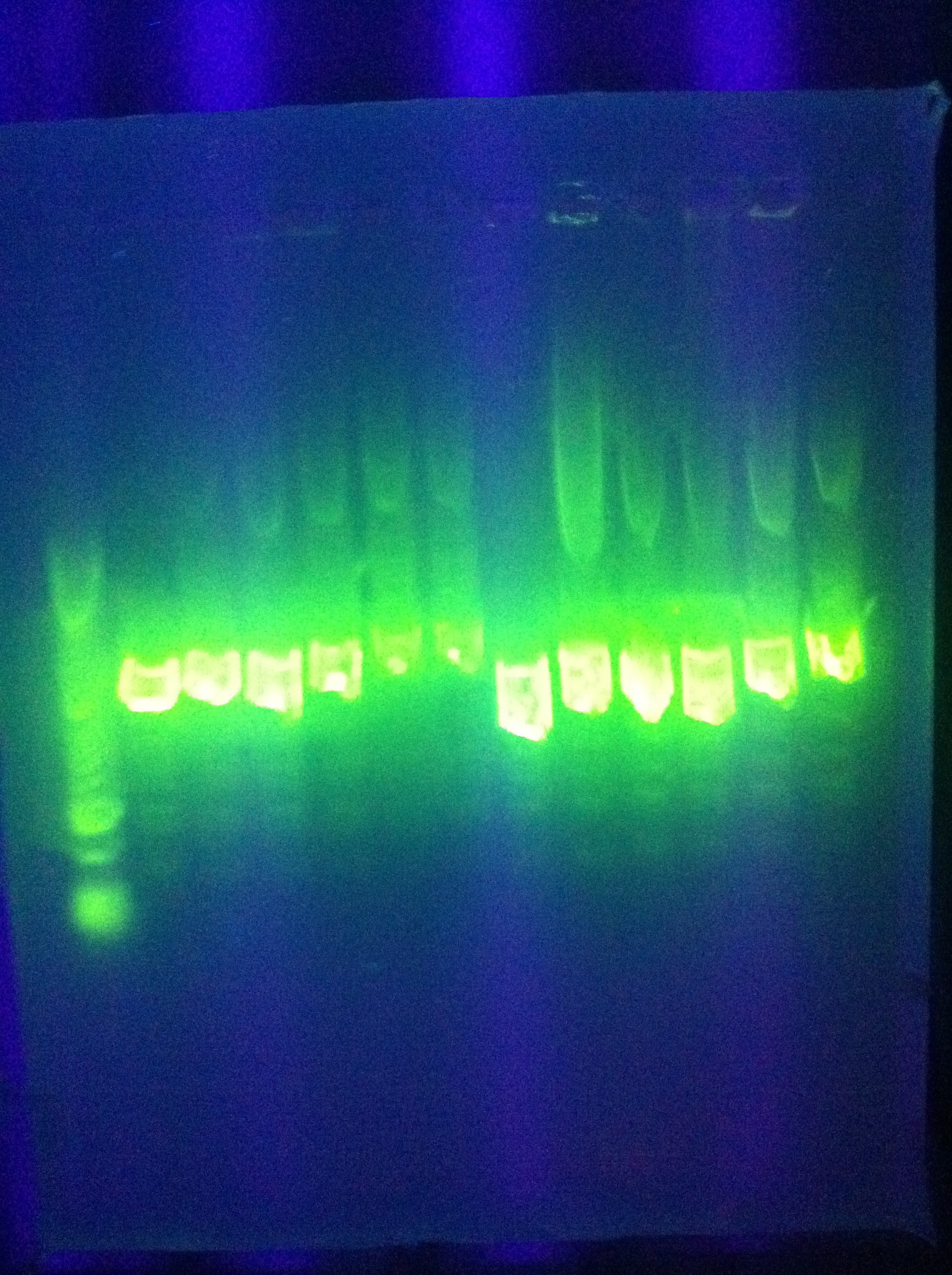
| + | * Samples (Primer): Result Summary | ||
| + | <br> [[Image:ASU_30_IMG_4171.jpg|400px]] | ||
| + | :* L2-7 (M13 F -20): + match for Seq1 and E0840 spacer present | ||
| + | :* L2-7 (M13 R -27): + match for Seq1 and E0840 spacer present | ||
| + | :* GFP in pRSF (T7 promoter): No substantial match | ||
| + | :* GFP in pRSF (T7 terminator): Several hundred identities for E0840, + match, e value = 0 | ||
| + | :* L2-6 (M13 F -20): No seq1 | ||
| + | :* L2-8 (M13 F -20): No seq1 | ||
| + | * Note: more samples were sequenced, available on DNASU. Samples compare to subject "Backbone and Cas Database" using blastn. | ||
}} <!-- close content attribute for menubar --> | }} <!-- close content attribute for menubar --> | ||
Latest revision as of 03:06, 29 September 2011
|
|
Monday, August 1
Tuesday, August 2
Wednesday, August 3
Thursday, August 4
Friday, August 5
Monday, August 8GROUP MEETING
Tuesday, August 9
TO DO for 8-10-11:
Wednesday, August 10
Thursday, August 11
Friday, August 12
Saturday, August 13
Sunday, August 14
Monday, August 15Team meeting
Tuesday, August 16
Wednesday, August 17
Thursday, August 18
Friday, August 19
Saturday, August 20
Incubated for 2 hours at 37 degrees.
Wednesday, August 24
Thursday, August 25
Friday, August 26
Saturday, August 27
Sunday, August 28
Wednesday, August 31
|
 "
"