Team:Arizona State/Notebook/July
From 2011.igem.org
(Difference between revisions)
Ethan ward (Talk | contribs) (Replaced content with "{{:Team:Arizona_State/Templates/sidebar|title=Notebook: July}}") |
|||
| (18 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{:Team:Arizona_State/Templates/ | + | __NOTOC__ |
| + | {{:Team:Arizona_State/Templates/main|title=Notebook: July|content= | ||
| + | | ||
| + | == Friday, July 1 == | ||
| + | * Ran a gel of PCRed CAS genes from last night | ||
| + | :* A1, B2, C3, D4, E5, F6, ladder 1; 10 uL of each sample placed in wells | ||
| + | * Conducted a ligation | ||
| + | * Transformation of NEB cells: | ||
| + | :* DA, double plated on LB+Kan plates | ||
| + | :* DB, double plated on LB+Kan plates | ||
| + | :* Seq1, double plated on LB+Kan plates | ||
| + | :* CMR, double plated on LB+Kan plates | ||
| + | == Saturday, July 2 == | ||
| + | * Plates from yesterday: colonies visible on all plates except CMR | ||
| + | :* Made overnight cultures of colonies | ||
| + | * Conducted ligation + transformation of RA, RB onto LB kan plates | ||
| + | * Ran gel of more pcr products | ||
| + | :* Big gel: c1, c2, c3, c4, c5, a1, a2, a3 | ||
| + | :* Small gel: e1, e2, e3, e4, e5, e6, f1, f2, f3, f4, f5, f6 | ||
| + | ::* No significant or usable bands visible on gels | ||
| + | == Tuesday, July 5 == | ||
| + | * Another PCR attempt at Cas genes, 2nd attempt with new primers: | ||
| + | :* A, C: didn't work | ||
| + | :* Attempted gel extraction | ||
| + | * Conducted miniprep | ||
| + | * Conducted restriction: | ||
| + | :* Seq1, DA, RA (ES, EX) in PSB1A3 | ||
| + | * Conducted ligation attempt to make tandem RSR, RS, and SR constructs: | ||
| + | :* Seq1 + Seq1, DA + DA, RA + RA; all into pSB1K3 | ||
| + | :* Transformation of ligation products, plated on LB+Kan (NEB cells) | ||
| + | * Also, another conducted ligation: | ||
| + | :* Seq1 (ES, XP) + PSB1A3 (EP) | ||
| + | :* Transformation, plated on LB+Amp (NEB cells) | ||
| + | * Made glycerol stocks of Seq1, DA, DB, RA, RB in PSB1K3 | ||
| + | == Wednesday, July 6 == | ||
| + | * Observed transformation results: | ||
| + | :* Kan plates: | ||
| + | ::* All successful (?) (Seq1 + Seq1, DA + DA, RA + RA), will be sequenced (ordered biobrick forward / reverse primers) | ||
| + | :::* Made liquid culture for miniprep tomorrow | ||
| + | :* Amp plates: | ||
| + | ::* No successes | ||
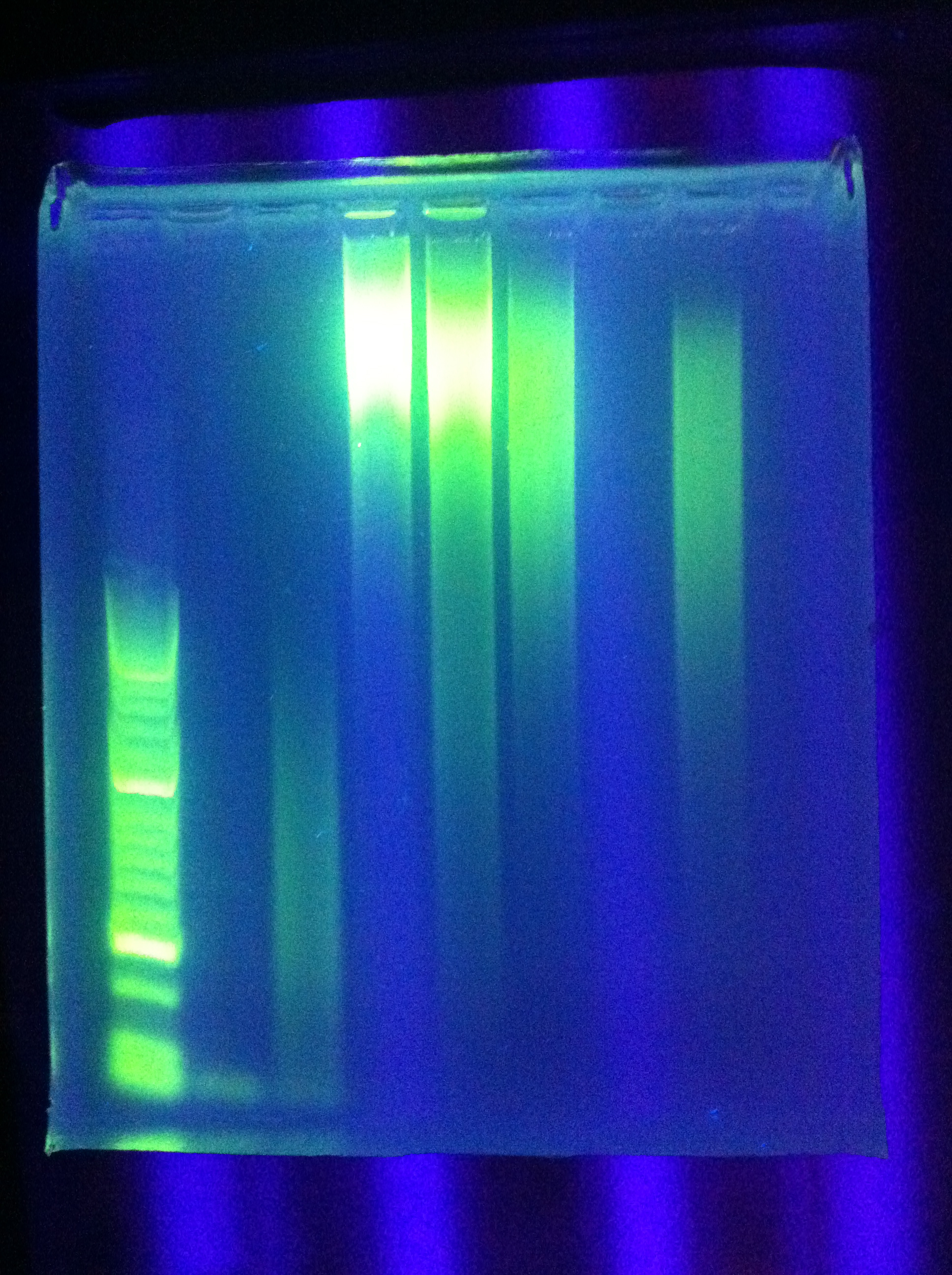
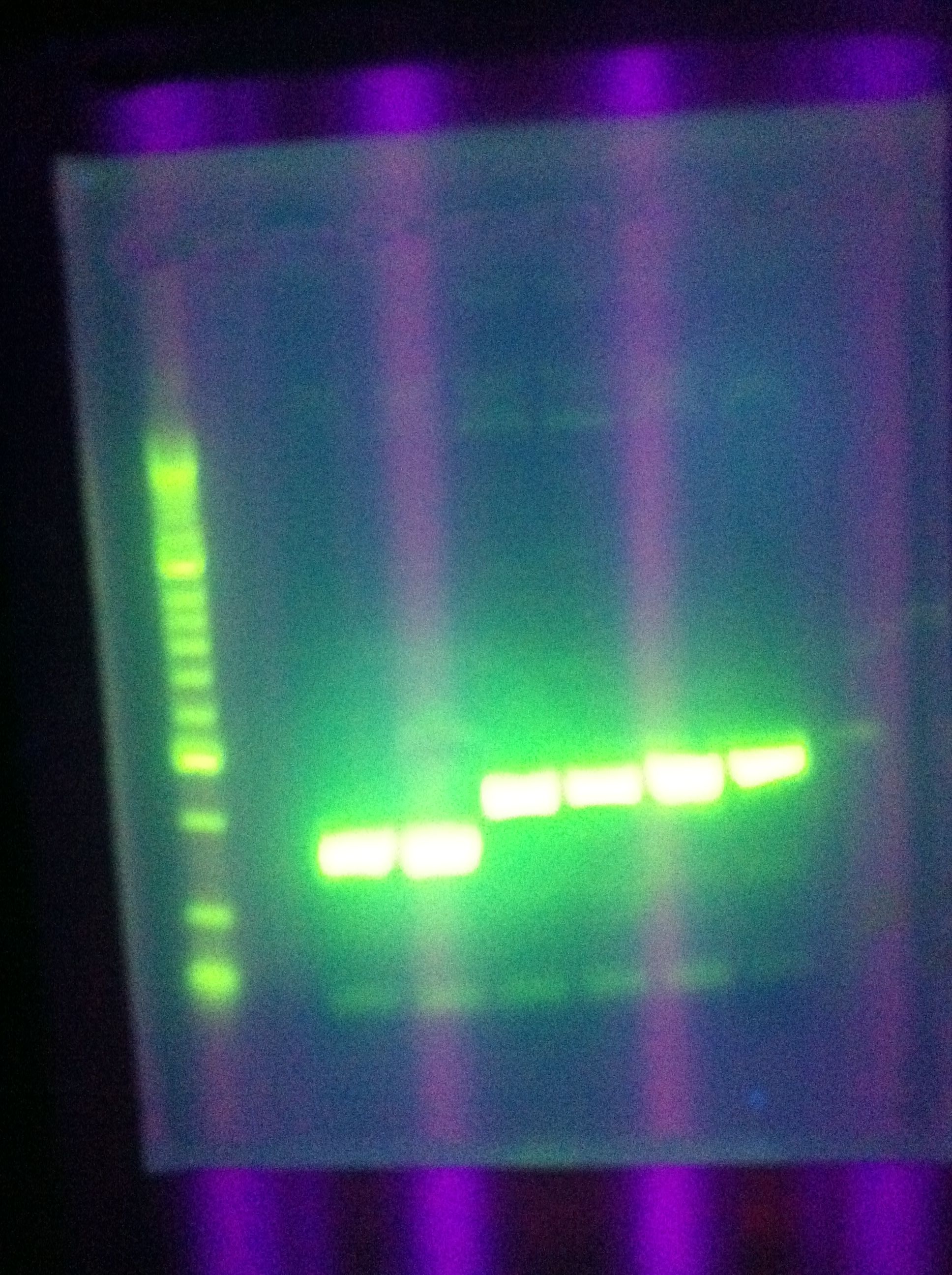
| + | * PCR: | ||
| + | :* Cas_b, Cas_c both unsuccessful | ||
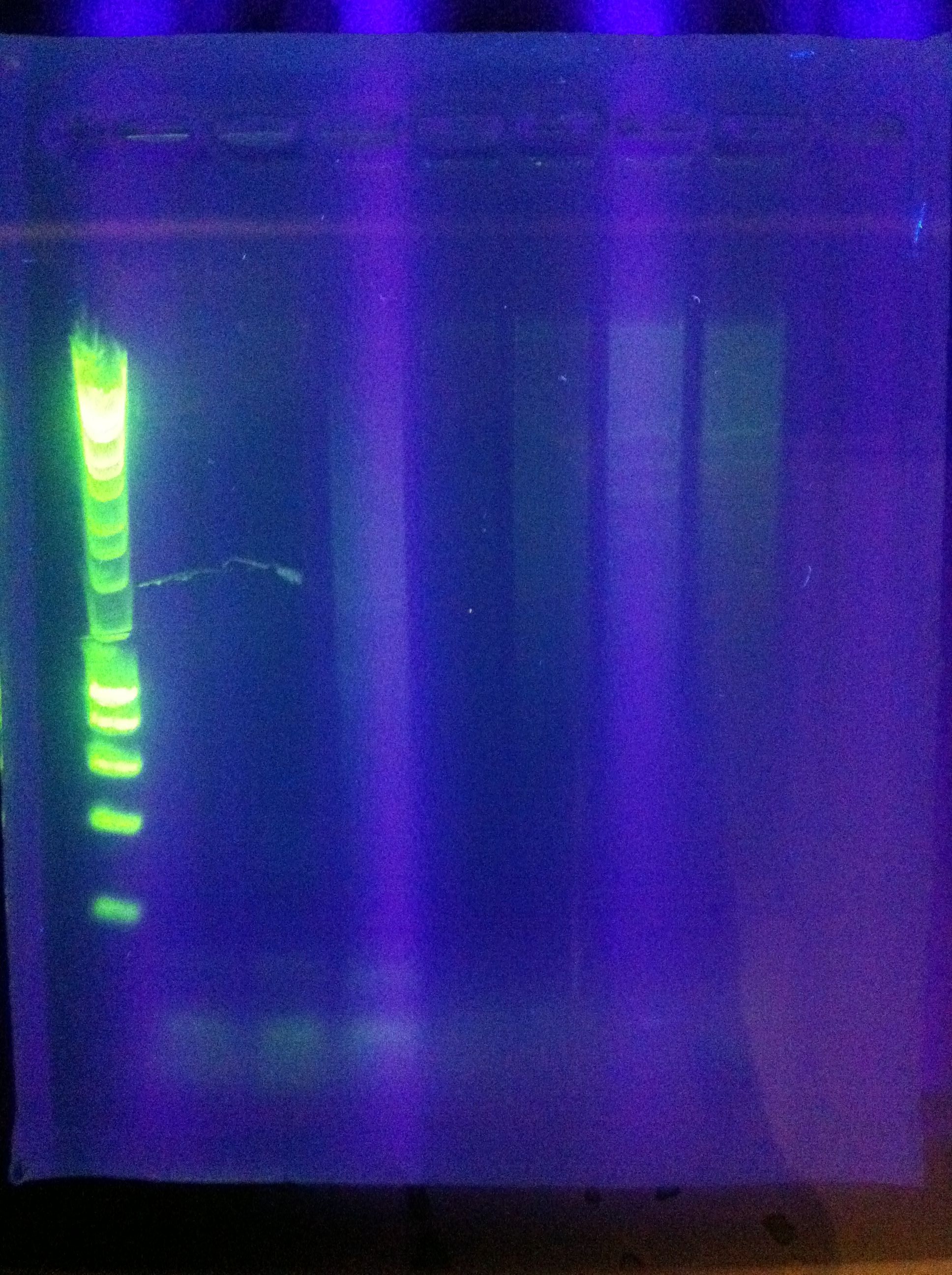
| + | <br> [[Image:ASU_76_gel1.jpg|400px]] | ||
| + | <br> [[Image:ASU_76_gel2.jpg|400px]] | ||
| + | :* Ran Cas_d on gel, will visualize tomorrow morning | ||
| + | :* Adjusted settings for cas_e_f, cas_3_r: 2 parallel runs on thermocycler | ||
| + | ::* touchdown to 70 using builtin touchdown | ||
| + | ::* constant 70 | ||
| + | * Primer design - do we need new primers??? | ||
| + | :* Is there melting temp mismatch? | ||
| + | * More lab supplies ordered (#4) | ||
| + | * Made new amp plates | ||
| + | == Thursday, July 7 == | ||
| + | * Miniprep of: | ||
| + | :* DADA (kit buffer) | ||
| + | :* RARA (kit buffer) | ||
| + | :* Seq1Seq1 (kit buffer, dan buffer) all in PSB1K3 | ||
| + | * Consensus: Dan's buffer that he created for lysis works | ||
| + | * Nanodrop results: (DADA 48 ng/uL, RARA 68 ng/uL, Seq1Seq1 Dan 73.5 ng/uL, Seq1Seq1 Kit 98 ng/uL) | ||
| + | * Glycerol stock of DADA, RARA, Seq1Seq1 all in PSB1K3 | ||
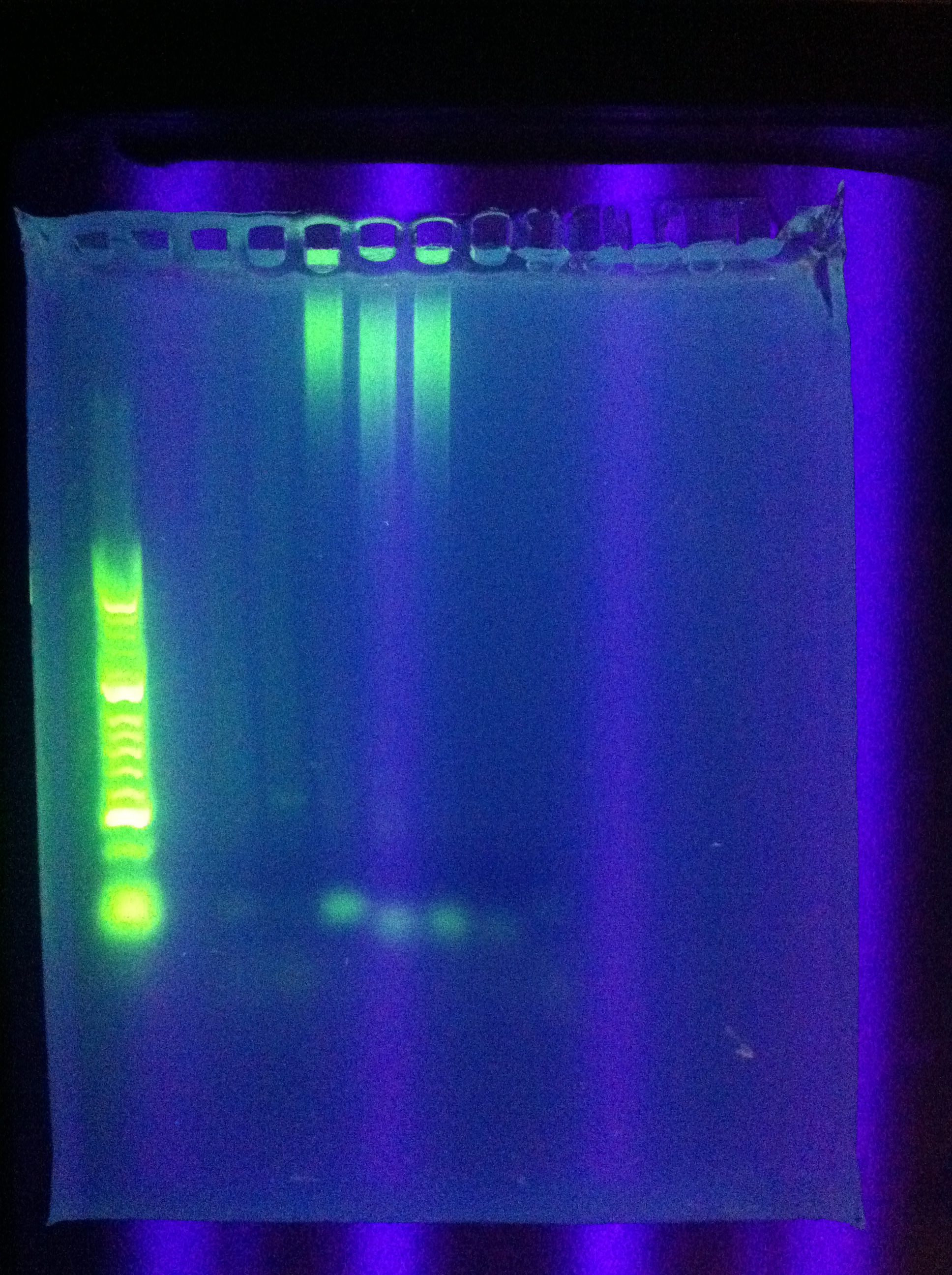
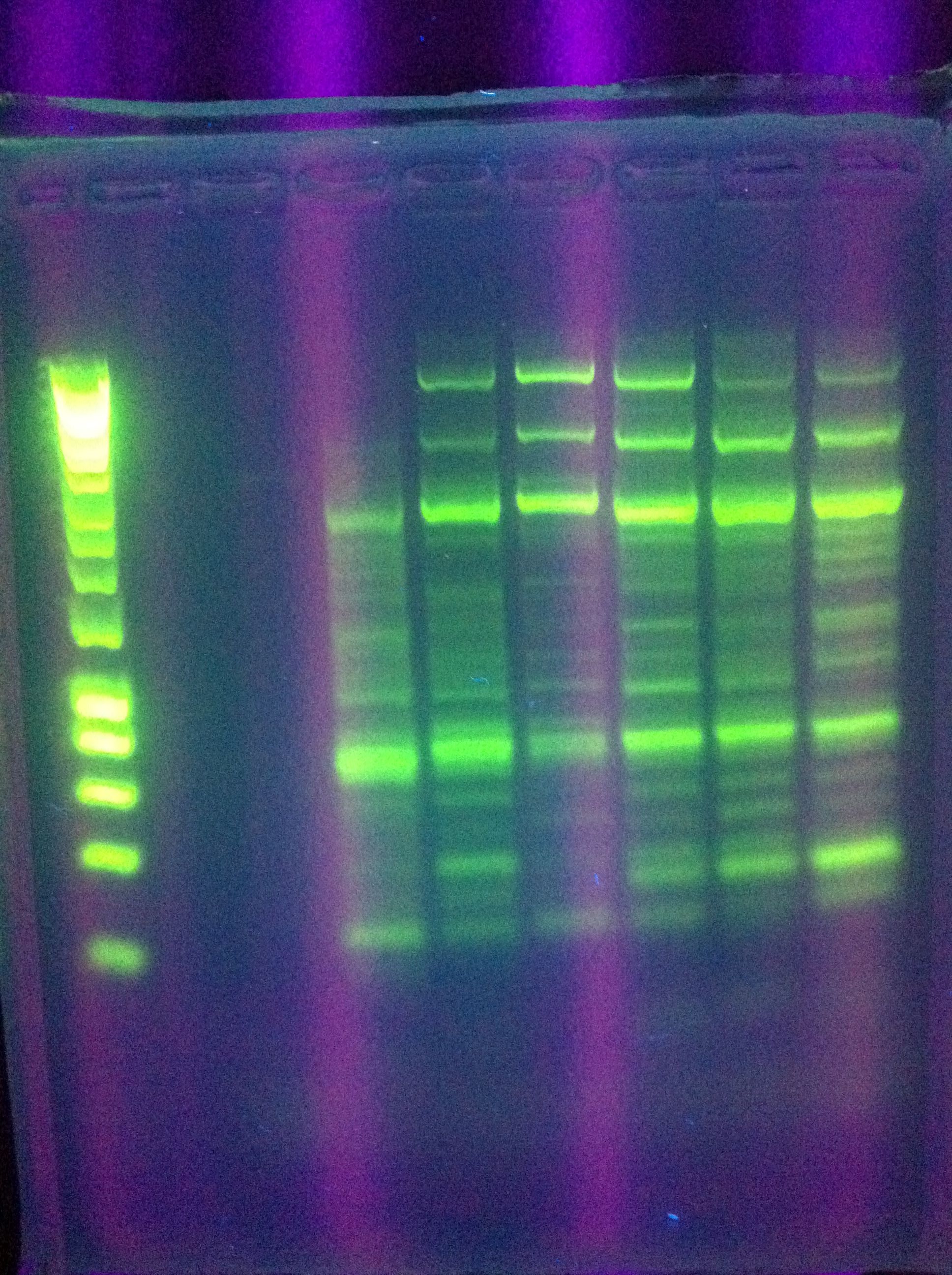
| + | * Gel of "homerun" attempt from yesterday, Block A touchdown + Block B constant 70 degC | ||
| + | <br> [[Image:ASU_77_mega_gel.jpg|400px]] | ||
| + | :* Unsuccessful, deformed gel near top, nonspecific binding, weird band at ~800bp | ||
| + | * Met w/ 3 grad students (Kurt, Jack, Bo) and had good discussion | ||
| + | :* Gel techniques: doing gels in 4 degree room, using X-tracta gel removers for gel band extraction | ||
| + | :* Assembly: problems with large insertions | ||
| + | :* Infusion (clonetech) | ||
| + | :* Transformation of biobrick promoter J23101 onto amp (2 plates) | ||
| + | * Streak plated E0840 GFP part onto amp from glycerol stock (3 plates) | ||
| + | * Determined theoretical 3 part PCR (casEDC, casBA, case), details in today's folder | ||
| + | * PCR attempt (see photo): | ||
| + | :* Block A - CasE forward, Cas3 reverse again but w/varying template DNA concentrations | ||
| + | ::* 225 ng/uL, 112.5 ng/uL, 56 ng/uL (previous starting conc. was 450, which is too high and could have caused problems) | ||
| + | ::* Settings stored as TCHDOWN (started @ 80deg, increment -.3) | ||
| + | :* Block B - Cas3 forward/reverse w/same varying template DNA concentrations | ||
| + | ::* Settings stored as IGEM707 | ||
| + | * Restriction: | ||
| + | :* Seq1Seq1 (ES, EX) | ||
| + | :* DADA (ES, EX) | ||
| + | :* RARA (ES, EX) | ||
| + | * Ligation (protocol from OpenWetWare): | ||
| + | :* 2x Seq1Seq1 | ||
| + | :* 2x DADA | ||
| + | :* 2x RARA | ||
| + | ::* ((used 22.5 uL water, 15 uL insert, 5 uL vector, 5 uL T4 buffer, 2.5 uL T4 ligase)) | ||
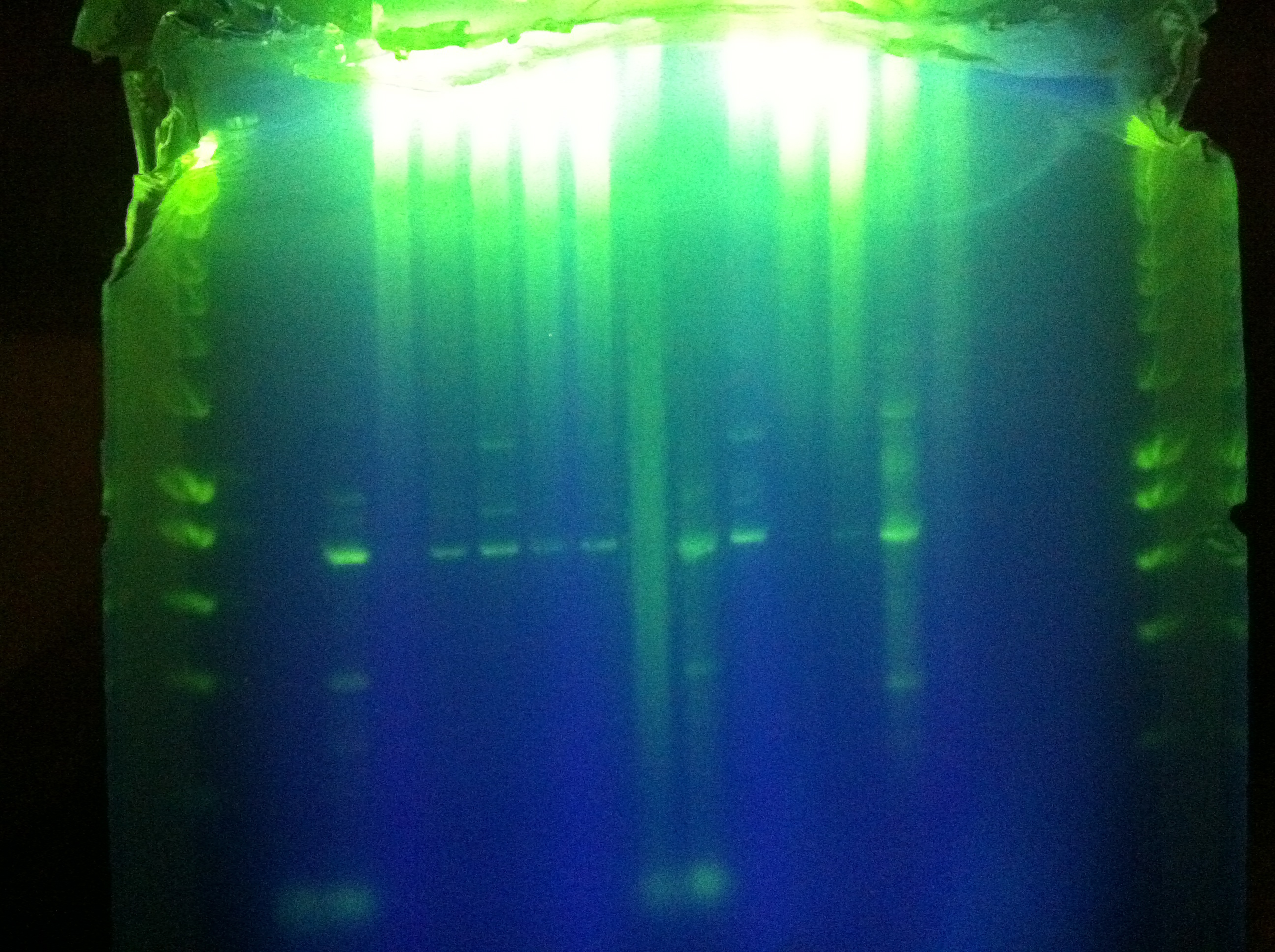
| + | :* Plated onto 3 kan plates using NEB protocol. | ||
| + | ::* 2 problems: | ||
| + | ::: 1. did not denature ligase before transformation | ||
| + | ::: 2. put cells in 37 room for 1 hour without adding SOC. SOC was added and the cells were shaken for another hour in the 37 room. | ||
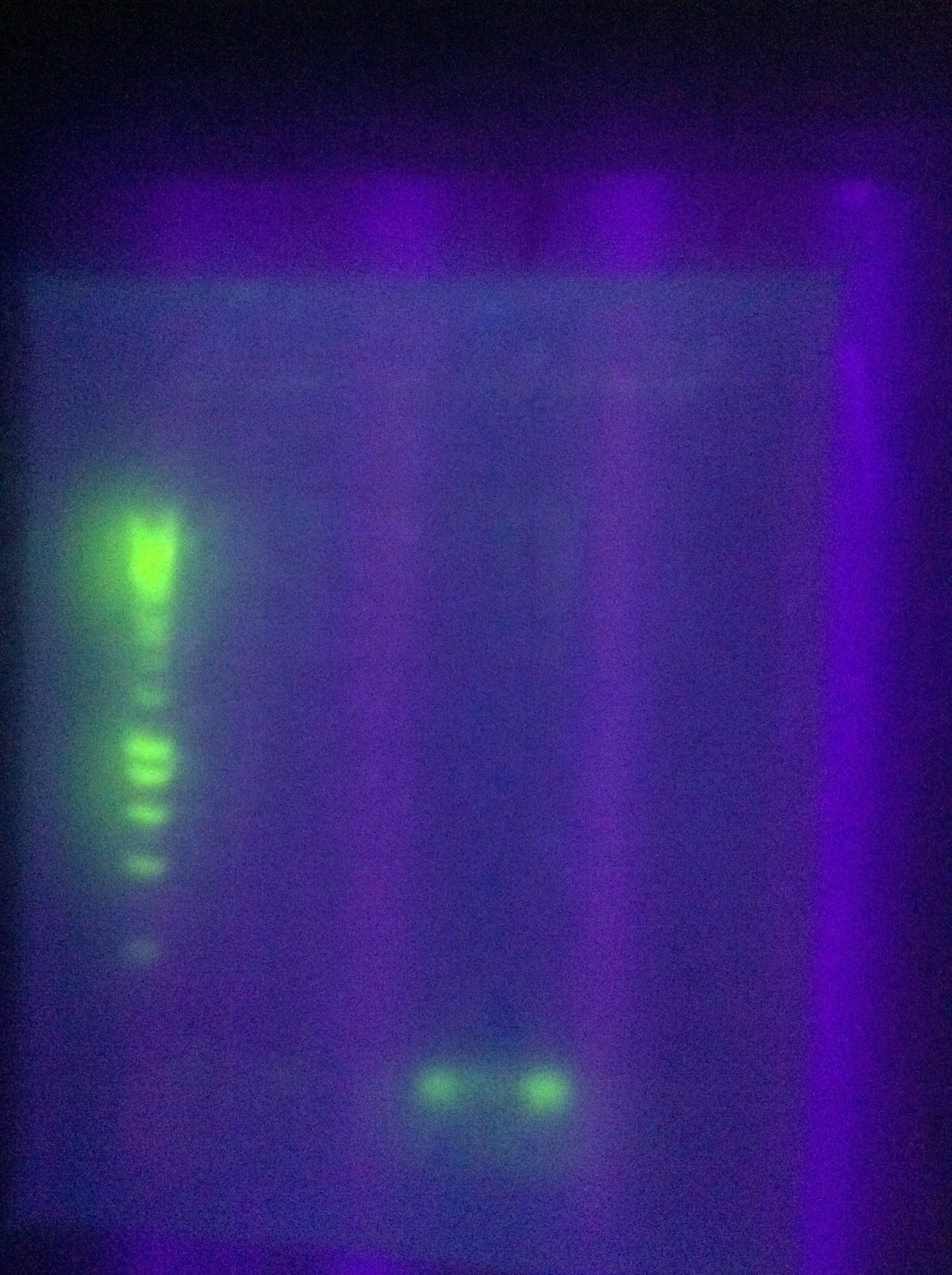
| + | <br> [[Image:ASU_77_plan1.jpg|400px]] | ||
| + | <br> [[Image:ASU_77_plan2.jpg|400px]] | ||
| + | == Friday, July 8 == | ||
| + | * GFP promoter transformation: | ||
| + | :* No success | ||
| + | * GFP streak plate: | ||
| + | :* Grew successfully | ||
| + | * DAx4, RAx4, Seq1 transformation: | ||
| + | :* Successful, but very small colonies | ||
| + | * Received order from iGEM (pSB1A3 and other agar stab) | ||
| + | :* transformed pSB1A3 plasmid to make stock | ||
| + | :* streak plated agar stab of J04430 | ||
| + | == Sunday, July 10 == | ||
| + | * Ran gel of array (1x, 4x): | ||
| + | :* Backbones visible for all | ||
| + | :* No visible inserts in plasmid backbone | ||
| + | * PCR of: | ||
| + | :* Homerun, EDC, AB, 3 | ||
| + | :* None of these attempts work | ||
| + | == Monday, July 11 == | ||
| + | * Transformation results: | ||
| + | :* pSB1A3: DNA contamination? Red colononies as well as normal, uncolored colonies growing on plates | ||
| + | ::* Possible contamination? We will need to sequence when we get biobrick primers | ||
| + | :* Agar stab of part J04430: normal (glowing) | ||
| + | * We made liquid cultures of: | ||
| + | :* SEQ1, DA, RA (8x) in PSB1k3 | ||
| + | :* PSB1A3 (pink, white colonies) | ||
| + | :* J04430 | ||
| + | |||
| + | * Restriction: | ||
| + | :* Restarting from 1x in PSB1K3 | ||
| + | :* Ginkgo: | ||
| + | ::* Seq1: ES, XP | ||
| + | ::* DA: ES, XP | ||
| + | ::* RA: ES, XP | ||
| + | ::* E0840: EP to get PSB1A3 backbone | ||
| + | :* Normal: | ||
| + | ::* Seq1: ES, EX | ||
| + | ::* DA: ES, EX | ||
| + | ::* RA: ES, EX | ||
| + | * ligation: | ||
| + | :* 2x Seq1, DA, RA 2 ways: | ||
| + | ::* pSB1K3 | ||
| + | ::* pSB1A3 | ||
| + | |||
| + | * Transformation of 14 plates: | ||
| + | :* New promoter (g18 on plate 1) in duplicate | ||
| + | :* Ligation products in duplicate | ||
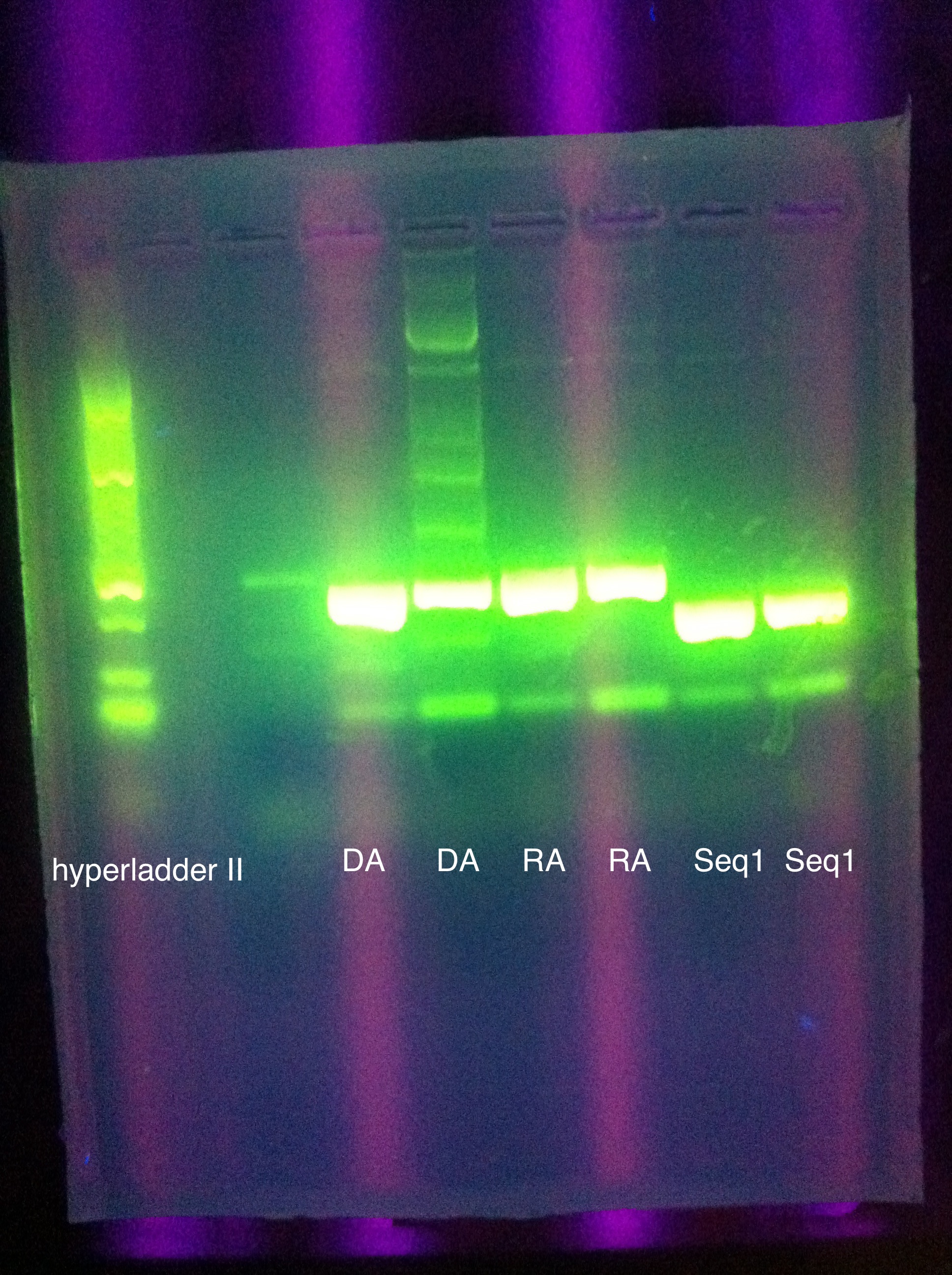
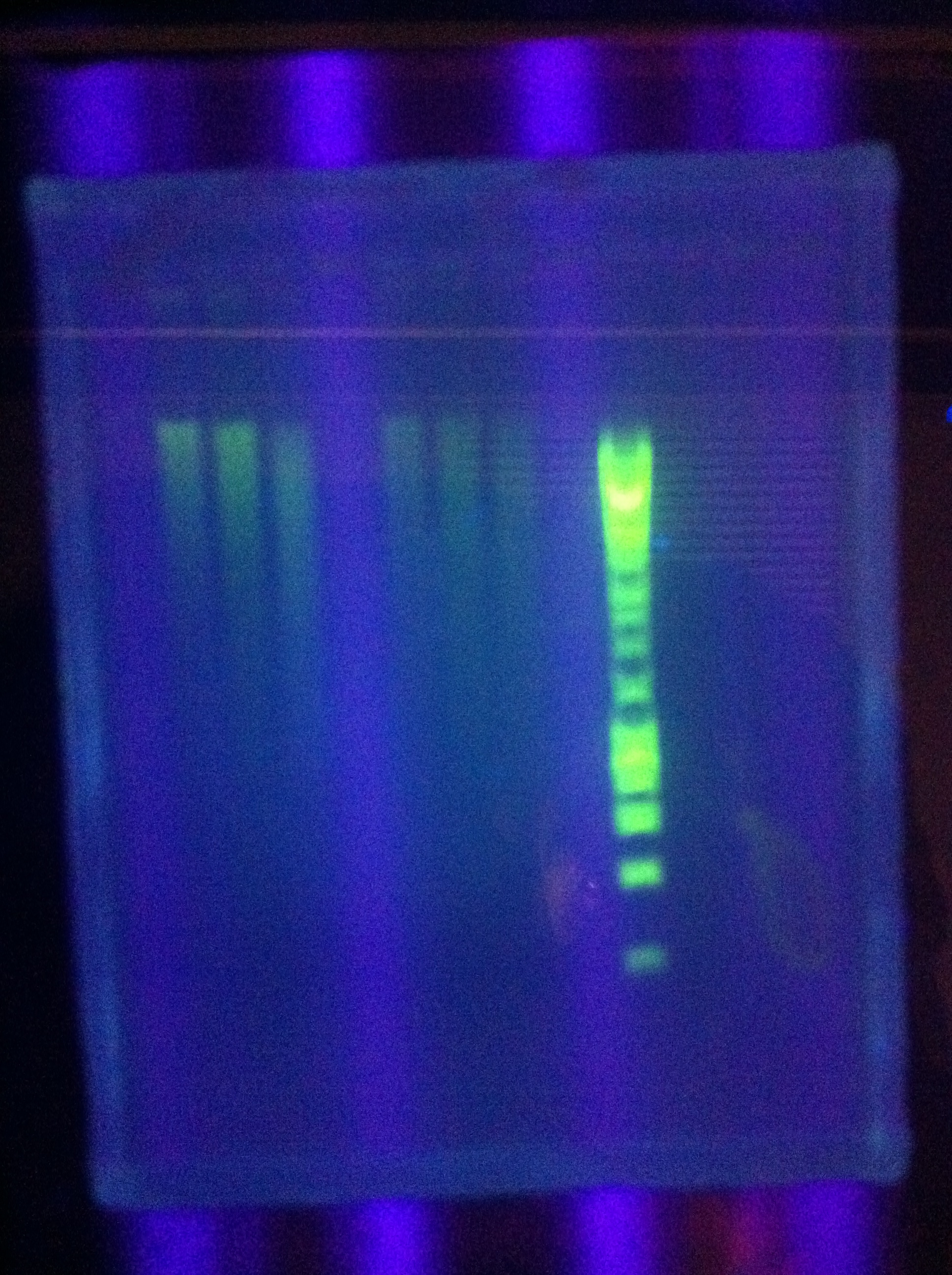
| + | <br> [[Image:ASU_711_8xDA.jpg|400px]] | ||
| + | <br> [[Image:ASU_711_gfp_construct_from_biobricks.jpg|400px]] | ||
| + | <br> [[Image:ASU_711_psb1a3_2.jpg|400px]] | ||
| + | * Planning: | ||
| + | :* Planned assembly of plasmid construct (cas genes, leader sequence, array) using pRSF-Duet plasmid | ||
| + | <br> [[Image:ASU_711_plan2.jpg|400px]] | ||
| + | == Tuesday, July 12 == | ||
| + | * transformation results: all successful | ||
| + | : 1. 2x seq1 normal in psb1a3 | ||
| + | : 2. 2x seq1 gingko | ||
| + | : 3. 2x ra normal in psb1a3 | ||
| + | : 4. 2x ra gingko in psb1k3 | ||
| + | : 5. 2x da normal in psb1a3 | ||
| + | : 6. 2x da gingko in psb1k3 | ||
| + | : 7. 2x seq1 normal in psb1a3 | ||
| + | : 8. 2x seq1 gingko in psb1k3 | ||
| + | : 9. 2x ra normal in psb1a3 | ||
| + | : 10. 2x ra gingko in psb1k3 | ||
| + | : 11. 2x da normal in psb1a3 | ||
| + | : 12. 2x da gingko in psb1k3 | ||
| + | : 13. promoter in psb1a3 | ||
| + | : 14. promoter in psb1a3 | ||
| + | * liquid cultures of: | ||
| + | :* promoter | ||
| + | :* e0840 | ||
| + | :* 2x seq1, da, ra | ||
| + | * miniprep using new ethanol protocol: | ||
| + | :* 8x (ra, da, seq1) | ||
| + | ::* nanodropped | ||
| + | ::* yields: 50-1000 ng / ul | ||
| + | :* psb1a3 red colony | ||
| + | * sequencing orders: | ||
| + | :* 1x, 2x, 4x (3 times each) | ||
| + | * submitted primers / synthesis order: | ||
| + | :* new cmr | ||
| + | :* new cas3 | ||
| + | :* new casabcde | ||
| + | :* adapter plasmids for leader sequences for cmr, cas | ||
| + | * ordered new restriction enzymes | ||
| + | <br> [[Image:ASU_712_progress.jpg|400px]] | ||
| + | == Wednesday, July 13 == | ||
| + | * We haven't been using alkaline phosphatase | ||
| + | :* Haven't been preventing vector self ligation | ||
| + | ::* We have nothing! probably | ||
| + | * ordered: | ||
| + | :* AAP | ||
| + | * Answered safety questions | ||
| + | * Miniprepped promoter | ||
| + | * Still waiting on sequencing results | ||
| + | == Thursday, July 14 == | ||
| + | * Miniprep: | ||
| + | :* All constructs in puc57 | ||
| + | * Restriction: | ||
| + | :* seq1: es, xp | ||
| + | :* da: es, xp | ||
| + | :* ra: es, xp | ||
| + | :* psb1K3: ep | ||
| + | :* promoter: sp | ||
| + | :* e0840: xp | ||
| + | * Ligation: | ||
| + | :* da + da | ||
| + | :* ra + ra | ||
| + | :* seq1 + seq1 | ||
| + | :* e0840 + promoter | ||
| + | * Transformation: | ||
| + | :* 2x da | ||
| + | :* 2x ra | ||
| + | :* 2x seq1 | ||
| + | :* E0840 + GFP promoter | ||
| + | == Friday, July 15 == | ||
| + | * Liquid cultures: | ||
| + | :* LB + Kan: | ||
| + | ::* 2x Seq1, DA, RA | ||
| + | * Transformation: | ||
| + | :* Seq1, ra, da in puc57 (original from biobasic) | ||
| + | * Sequencing results: 3: | ||
| + | :* Did not show inserts we want | ||
| + | :* A piece of puc57, E. Coli genome was sequenced | ||
| + | ::* Possible contamination? | ||
| + | :* Use of AAP will prevent this in the future | ||
| + | * Fixed and submitted IDT leader sequence order | ||
| + | * Restriction/ligation of: | ||
| + | :* E0840, GFP promoter | ||
| + | :* normal (SP, XP): | ||
| + | ::* used AAP on both E0840 (XP) and promoter (SP) | ||
| + | ::* EP vector (kan) was added that should not have been added | ||
| + | ::* Was subsequently transformed onto kan plate, should have been amp | ||
| + | :* Ginkgo (ES, XP, EP): | ||
| + | ::* Used AAP on psb1k3 vector (EP) | ||
| + | ::* Transformed and grown on kan plates | ||
| + | * Primers arrived! | ||
| + | * PCR | ||
| + | :* CasABCDE | ||
| + | ::* Template concentration gradient (225 ng/uL, 112.5 ng/uL, 56 ng/uL) | ||
| + | ::* For each concentration, mgcl2 gradient (null, 1, 2, 4, 8) | ||
| + | :* Cas3 | ||
| + | ::* Template concentration gradient (225 ng/uL, 112.5 ng/uL, 56 ng/uL) | ||
| + | ::* For each concentration, mgcl2 gradient (null, 1, 2, 4, 8) | ||
| + | == Saturday, July 16 == | ||
| + | * Check transformants | ||
| + | :* GFP construct (e0840, promoter) did not grow 3: | ||
| + | :* 2x seq1, da, ra in psb1k3 | ||
| + | * Liquid cultures | ||
| + | :* Seq1, DA, RA in puc57, LB+Amp | ||
| + | :* Original k12, LB | ||
| + | * Miniprep | ||
| + | :* 2x seq1, da, ra in psb1k3 | ||
| + | :* Kylie will nanodrop this afternoon | ||
| + | * Glycerol stock | ||
| + | :* 2x seq1, da, ra in psb1k3 | ||
| + | :* Threw away glycerol stocks of old 8x, 4x, 2x | ||
| + | * Gel electrophoresis | ||
| + | :* CasABCDE | ||
| + | ::* Small gel, absolutely nothing visible | ||
| + | :* Cas3 | ||
| + | ::* Big gel, also absolutely nothing 3: | ||
| + | :* CMR | ||
| + | ::* SUCCESS - CMR is the thermocycler favorite | ||
| + | <br> [[Image:ASU_716_cmr_success.jpg|400px]] | ||
| + | ::* Extracted by Ethan | ||
| + | ::* This means our polymerase is fine, probably the template k12 that was wack | ||
| + | * Potential issues: | ||
| + | :* Template DNA (which is why we are doing a new liquid culture of k12 for new genome prep) | ||
| + | :* Polymerase problems? | ||
| + | :* Something w/ master mix? | ||
| + | :* Trinette the PCR machine didn't like the train of thermocycles running on her all day | ||
| + | ::*Choo Choo! | ||
| + | :* PCR gnomes (DNA trafficking) | ||
| + | * PCR: | ||
| + | :* CMR (using new primers) | ||
| + | :* 220ng/ul null, 1, 2, 4, 8, 10 | ||
| + | :* 181ng/ul 1, 2, 4, 10 | ||
| + | == Sunday, July 17 == | ||
| + | * Did a genome extraction of K-12 | ||
| + | * Miniprep: | ||
| + | :* Seq1, ra, da in puc57 | ||
| + | :* Very low concentrations likely due to inclusion of precipitate -- be careful w/ this step! | ||
| + | * Restriction: | ||
| + | :* promoter: sp + aap | ||
| + | :* e0840: xp + aap (accident) | ||
| + | :* 2x seq1: es xp + aap (accident) | ||
| + | :* 2x ra: es xp | ||
| + | :* 2x da: es xp | ||
| + | :* psb1a3: ep + aap | ||
| + | * Ligation: | ||
| + | :* promoter + e0840 | ||
| + | :* 2x seq1 -> 4x | ||
| + | :* 2x ra -> 4x | ||
| + | :* 2x da -> 4x | ||
| + | * Transformation: | ||
| + | :* Using CCMB80 cells (old) | ||
| + | ::* old cells may not be competent / alive | ||
| + | * PCR: | ||
| + | :* ABCDE: | ||
| + | ::* start 70, -0.2 / cycle | ||
| + | ::* final: 64 | ||
| + | :* CAS3 | ||
| + | ::* start 63, -0.2 / cycle | ||
| + | ::* final: 57 | ||
| + | :* gel results: | ||
| + | ::* ABCDE: very faint band | ||
| + | ::* cas3: no bands | ||
| + | <br> [[Image:ASU_717_cas_gel.jpg|400px]] | ||
| + | == Monday, July 18 == | ||
| + | * Made new TSS cells | ||
| + | * Met with Catherine, got some good resources for modeling | ||
| + | :* She will be less busy in August and can help us further | ||
| + | * PCR: | ||
| + | :* CasABCDE using a lower starting temp by 5 degrees, still touchdown | ||
| + | :* Gel results: no bands at all | ||
| + | * RLT: | ||
| + | :* Traditional GFP construct | ||
| + | ::* GFP promoter (SP + AAP) | ||
| + | ::* E0840 (XP + gel extract) | ||
| + | :* Ginkgo GFP construct | ||
| + | ::* GFP promoter (ES) | ||
| + | ::* E0840 (XP) | ||
| + | ::* psb1k3 (EP + aap) | ||
| + | :* 4x constructs (ligation) | ||
| + | ::* seq1: ex, xp | ||
| + | ::* da: ex, xp | ||
| + | ::* ra: ex, xp | ||
| + | ::* psb1a3: ep + aap | ||
| + | * submitted sequencing: | ||
| + | :* 2x: da, ra, seq1 | ||
| + | :* CMR (f, r) | ||
| + | * Nanodropped 1x in puc57 miniprep by Madeline | ||
| + | :* Horrible results! | ||
| + | :* Nisarg redid this miniprep today, will nanodrop tomorrow | ||
| + | ::* ((update: much better!)) | ||
| + | * Made TSS cells w/ old BL21 DE3 | ||
| + | :* We have asked Jon for fresh cells to make better comp cells | ||
| + | == Tuesday, July 19 == | ||
| + | * Transformation results: | ||
| + | :* 4x Seq1 successful | ||
| + | ::* However, we know this isn't actually 4x Seq1 | ||
| + | :* 4x DA, RA unsuccessful | ||
| + | :* GFP construct unsuccessful | ||
| + | * Sequencing results: | ||
| + | :* CMR: good =3 | ||
| + | :* All else: nothing that we need 3: | ||
| + | ::* Just vector sequences | ||
| + | * New sequencing order: | ||
| + | :* Putative RFP | ||
| + | :* Original biobasic synthesis products in puc57 (with new puc57 primers) | ||
| + | ::* da, ra, seq1 | ||
| + | :* Our 1x transformation of biobasic synthesis products in puc57 (with new puc57 primers) | ||
| + | ::* 1xda, 1xra, 1xseq1 | ||
| + | :* Genomic DNA from mg1655 (k-12) to see if they can pull out cas gene sequences that we can't | ||
| + | ::* abcde f/r and cas3 f/r | ||
| + | * PCR: | ||
| + | :* Block A: | ||
| + | ::* Settings we used to get faint bands | ||
| + | ::* CasABCDE | ||
| + | ::* Touchdown: 70, -0.2 / cycle -> 63 | ||
| + | :* Block B: | ||
| + | ::* Cas3 | ||
| + | ::* Gradient with 3 rows from 72 to 55 | ||
| + | * Late night gel results: (Madeline and Joseph) | ||
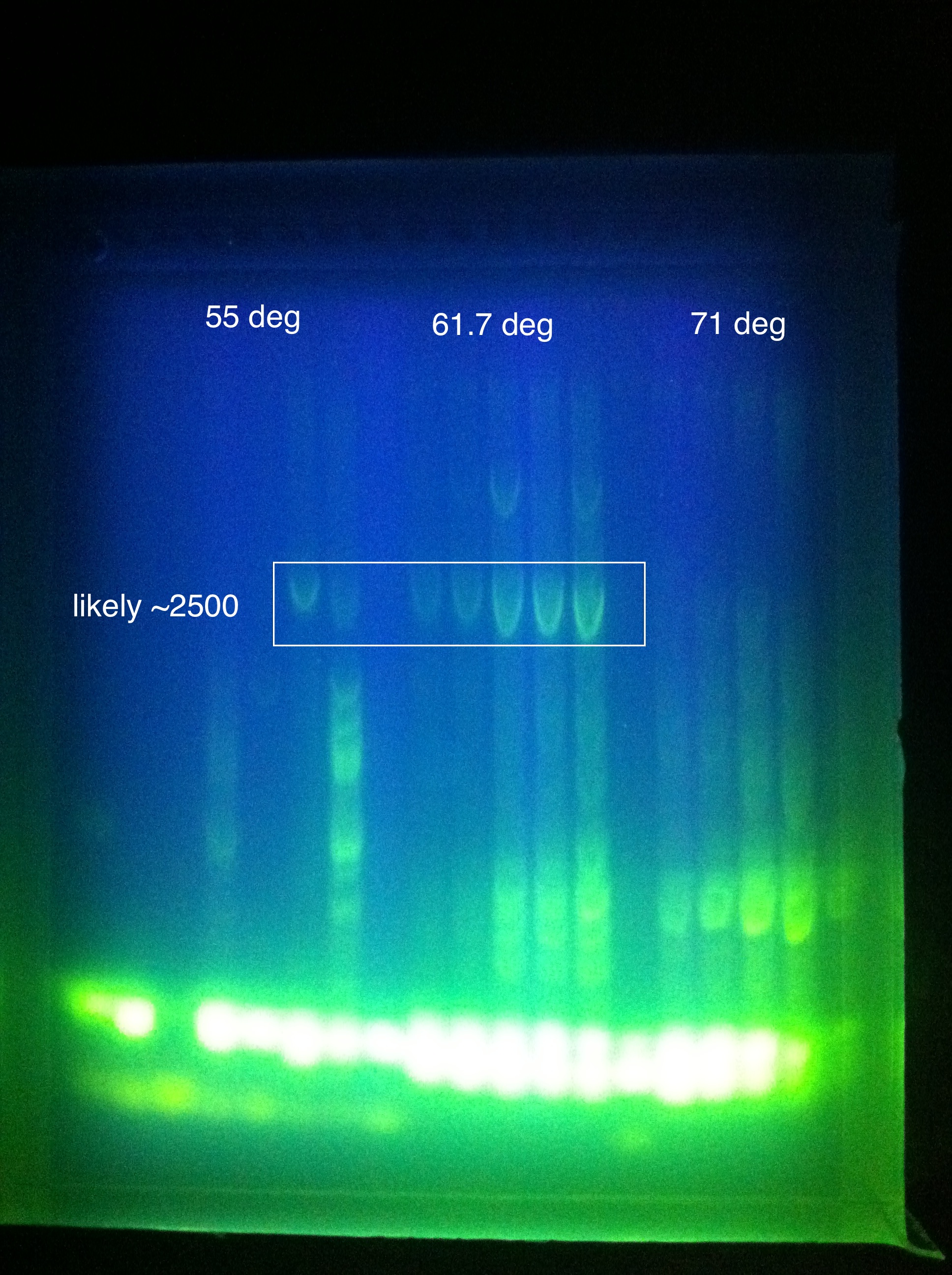
| + | <br> [[Image:ASU_719_casABCDE.jpg|400px]] | ||
| + | :* CasABCDE: broad bands, perhaps desired band at ~4300 faintly @ higher magnesium concentrations | ||
| + | <br> [[Image:ASU_719_cas3.jpg|400px]] | ||
| + | :* Cas3: broad bands, also no ladder | ||
| + | ::* So we THINK we got the desired band in the "midrange" temperature, which corresponds to "third up" row and relatively high magnesium chloride concentration | ||
| + | ::* SO we should do another PCR at approximately slightly above 60 somewhere | ||
| + | ::* Suggestion: gradient between 65 and 60 | ||
| + | ::* Also: what does it mean that it is curved? | ||
| + | ::* Also: what is the brightness at the bottom? happened in both gels... | ||
| + | * Restriction, Ligation and Transformation (RLT): | ||
| + | * Tried to gel E0840 cut w/ XP to get higher concentration of insert for ligation | ||
| + | ::* No bands at all 3: | ||
| + | :* Instead, ligated / transformed purified GFP insert from yesterday | ||
| + | :* @ 3ng/uL with approximately 40 uL, therefore ~120 ng of GFP insert (E0840) | ||
| + | ::* w/ 5 uL of vector (promoter cut at SP and w/ AAP) | ||
| + | ::* 5 uL of T4 ligase buffer | ||
| + | ::* 2.5 uL of T4 ligase | ||
| + | ::* total: 52.5 uL reaction | ||
| + | * Other news: | ||
| + | :* Got new bacterial strains from Jon | ||
| + | :* Submitted Barrett reimbursement forms for gas, food | ||
| + | :* Met w/ Alana Labelle about safety, who told us to complete a "Preliminary Hazard Assessment" form | ||
| + | :* New primers for duet vector and puc57 arrived | ||
| + | :* GOT ICE ACCESS!!! | ||
| + | :* Kylie/Keith battle has begun | ||
| + | :* Xiao is proud of us :3 | ||
| + | * To do: | ||
| + | :* Email iGEM to see what's up w/ Indianapolis accommodations | ||
| + | :* Fill out form | ||
| + | == Wednesday, July 20 == | ||
| + | * Transformation results: | ||
| + | :* 2 tiny colonies on one plate (GFP) | ||
| + | :* Made liquid culture (lb amp) | ||
| + | ::* Did not grow | ||
| + | * PCR: | ||
| + | :* ABCDE: starting: 71, -0.2/cycle -> 64 | ||
| + | :* Cas3: another gradient using tighter range around best temp from yesterday | ||
| + | ::* 65.6 | ||
| + | ::* 63.1 | ||
| + | ::* 61.2 | ||
| + | ::* 58.5 | ||
| + | :* Results: | ||
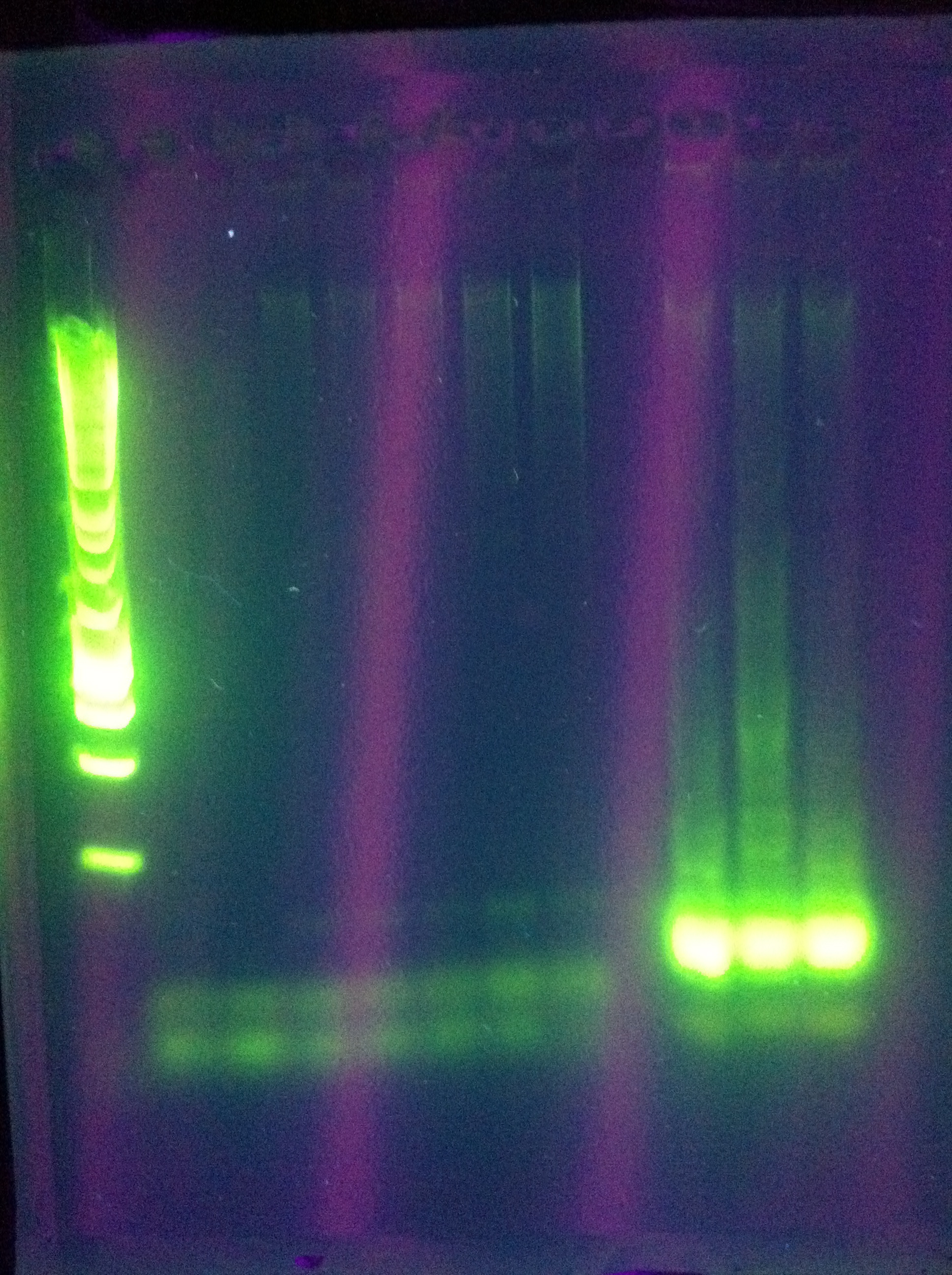
| + | <br> [[Image:ASU_720_CasABCDE.jpg|400px]] | ||
| + | ABCDE: nothing usable | ||
| + | <br> [[Image:ASU_720_Cas3.jpg|400px]] | ||
| + | ::* CAS3: small band at correct location | ||
| + | :* Need to order new primers with less dimerization potential! | ||
| + | == Thursday, July 21 == | ||
| + | * Heat shocked overnight RE digests (4 tubes of each for maximal gel results) | ||
| + | :* E0840 | ||
| + | :* Seq1, RA, DA | ||
| + | * Gel of RE results | ||
| + | :* Very clear bright bands for (ES) E0840 vector, insert | ||
| + | ::* Extracted insert | ||
| + | :* No visible insert for Seq1, but backbone visible | ||
| + | * PCR Insert Amplification, Attempt #1 | ||
| + | :* Taq polymerase from Bioline | ||
| + | :* Notable: 30 sec elongation time | ||
| + | * Gel of PCR Results | ||
| + | <br> [[Image:ASU_721_PCR_Insert_Amplification_Attempt_1.jpg|400px]] | ||
| + | :* faint bands at ~1000bp | ||
| + | :* likely due to 30sec elongation, will try again w/shorter time | ||
| + | * Late Night: | ||
| + | * Gel of Restricted (ES) DA, RA | ||
| + | <br> [[Image:ASU_721_PCR_Insert_Amplification_Attempt_2_(Seq1,_DA,_RA).jpeg|400px]] | ||
| + | :* 1% Agarose, 40mL total | ||
| + | :* Showed very strong, clear bands for backbone, no inserts | ||
| + | * PCR Insert Amplification, Attempt #2 | ||
| + | :* Used Phusion instead of Biolase product | ||
| + | :* Annealing temp: 58 deg, based off of NEB calculator as recommended by Phusion protocol | ||
| + | :* Annealing time: 10 sec (low end) | ||
| + | :* Elongation @ 72 degrees for 5 seconds (much lower than previous attempt, corresponds to 170-330 bp) | ||
| + | :* 25 total cycles (low end) | ||
| + | :* 5 minute extension (low end) | ||
| + | * Gel Result: | ||
| + | :* Very strong, medium thick bands | ||
| + | :* Seq1 ~ 130-180bp | ||
| + | :* DA/RA ~ 190-220bp | ||
| + | * Other notes: | ||
| + | :* Met with Kurt and a new grad student today at 11 as usual. We discussed various methods to improve our yields in assembling the RSR array, including using PCR amplification with the pUC57 primers we used for sequencing to create the insert in excess prior to gel extraction. Also discussed PCRing PCR products to increase yield (we will likely try this for CasABCDE and Cas3 w/ the current primers). | ||
| + | == Friday, July 22 == | ||
| + | * PCR Amplification of pUC57 inserts | ||
| + | :* 4 second elongation (instead of 5 sec), 35 cycles (instead of 25) | ||
| + | * Gel Results | ||
| + | <br> [[Image:ASU_722_Gel_of_PCR_amplification_of_inserts.jpg|400px]] | ||
| + | :* same major bands as first attempt (~150 for seq1, ~200 for DA, RA) | ||
| + | :* more minor bands than before | ||
| + | :* might be contamination in second DA well | ||
| + | * Try again | ||
| + | :* PCR with 4 second elongation, but 25 cycles | ||
| + | :* Will gel small amount and if only 1 band is present, will restrict directly | ||
| + | * Nanodropped | ||
| + | :* Seq1, RA from per amp + gel extraction (~22ng/uL, ~12ng/uL) | ||
| + | :* E0840 gel extraction (~6ng/uL) | ||
| + | * GFP Restriction, Ligation | ||
| + | :* Previously restricted (XP) E0840 insert from gel extraction | ||
| + | :* (SP) Promoter in pSB1A3 | ||
| + | :* RAGE, Seq1RE into pSB1A3 | ||
| + | ::* Problem: water bath was too hot (~65) and enzymes were denatured | ||
| + | ::* Will try again tomorrow | ||
| + | * Ordered new primers | ||
| + | :* CasABCDE & Cas3 | ||
| + | :* Much lower affinity for dimerization | ||
| + | :* Nice, close melting points | ||
| + | * Ordered Phusion, enzymes (SpeI, DpnI), PCR cleanup kit | ||
| + | * Sent in sequencing requests | ||
| + | * Seq1, RA gel extracts (Forward and reverse primers separately) | ||
| + | == Saturday, July 23 == | ||
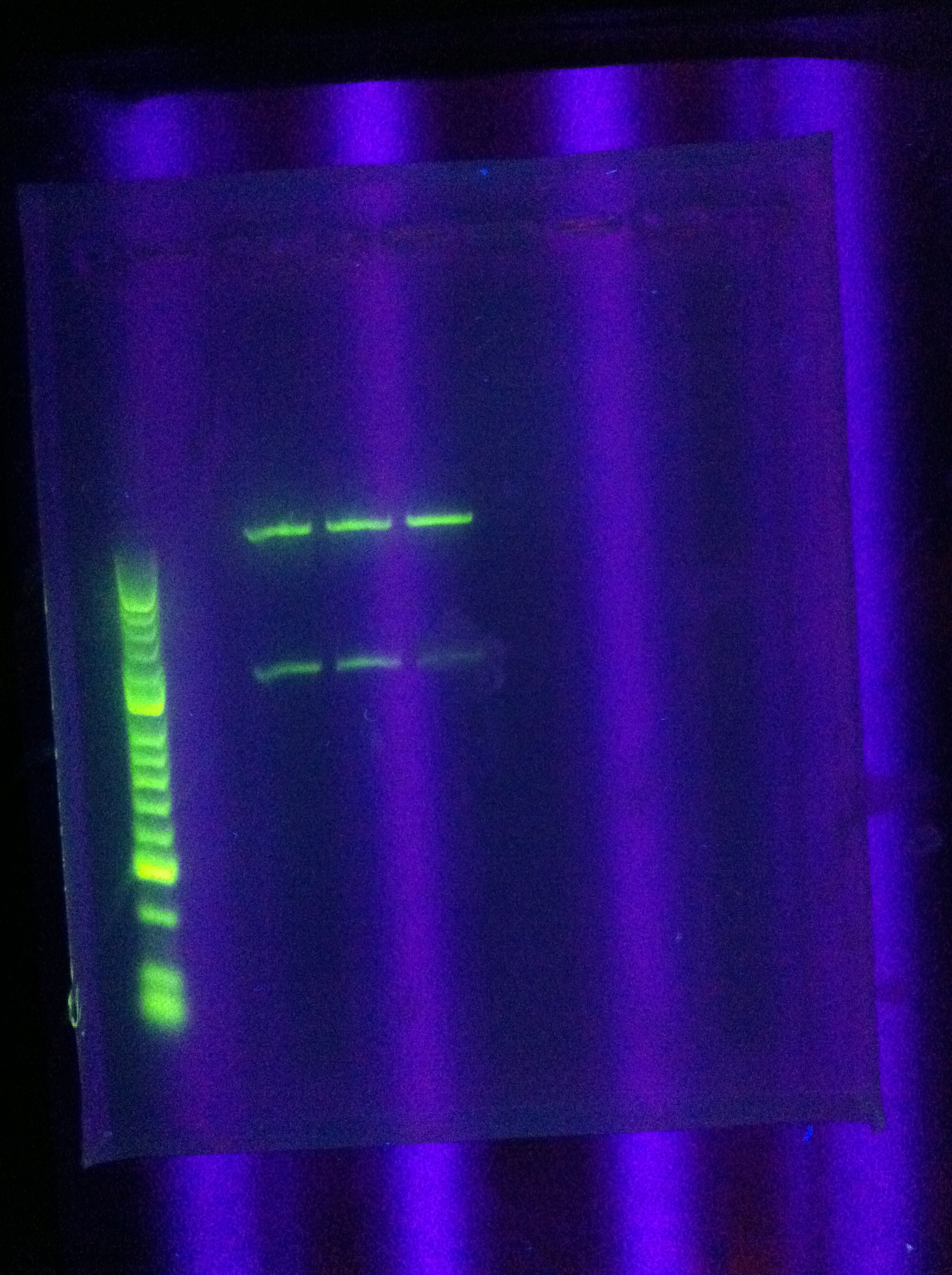
| + | * Ran gel of PCR amplification results | ||
| + | :* (ran out of Seq1, so RA and DA were gelled) | ||
| + | :* 10 uL in each well (2uL buffer, 2uL product, 6uL water) | ||
| + | * Results | ||
| + | <br> [[Image:ASU_723_Gel_of_PCR_amplified_DA,_RA.jpg|400px]] | ||
| + | :* very clear bands (except for DA w/4 mgcl2, which was faint/blurry) | ||
| + | * Restriction Digest | ||
| + | :* DA, RA from PCR amplification (no gel extraction) | ||
| + | :* E, P digest | ||
| + | :* Stored at -20 | ||
| + | * Check transformant from Friday | ||
| + | :* A single small colony visible on the GFP plate | ||
| + | :* No glow visible, but will keep growing it up in case | ||
| + | * Miniprep of Seq1, RA, DA in puc57 | ||
| + | :* added another round of centrifugation in step 11 | ||
| + | :* NANODROP RECORD!!! 3412 ng/uL by Joseph Flay | ||
| + | * Glycerol stocks of BL21 DE3, MG1655 | ||
| + | == Sunday, July 24 == | ||
| + | * Checked GFP transformation colony from Friday again | ||
| + | :* Single colony that hadn't glowed before was much bigger | ||
| + | :* IT GLOWS | ||
| + | : --> Liquid culture | ||
| + | :* PCR of CasABCDE, Cas3 with new settings | ||
| + | <br> [[Image:ASU_724_IMG_3932.jpg|400px]] | ||
| + | :* Preparation for extraction, "PCRception" (PCRing the PCR results) | ||
| + | :* Included an elongation step in each cycle, which was not done for the previous runs | ||
| + | :* Settings stored in PCR machine as ABCDE724, CAS3724 | ||
| + | * Restriction | ||
| + | :* RAGE (code for RA gel extraction), Seq1 GE (Seq1 gel extraction) (EP) | ||
| + | :* RFP in pSB1A3 (EP) + AAP | ||
| + | * Ligation | ||
| + | :* RAGE + pSB1A3 | ||
| + | :* Se1GE + pSB1A3 | ||
| + | :* PCR RA + pSB1A3 (messed this one up, probably won't grow) | ||
| + | :* PCR DA + pSB1A3 | ||
| + | * Transformation of ligation products | ||
| + | * Did dishes | ||
| + | == Monday, July 25 == | ||
| + | * Autoclaved and finished dishes | ||
| + | * Checked transformants | ||
| + | :* RAGE + pSB1A3 (growth) | ||
| + | :* Se1GE + pSB1A3 (growth) | ||
| + | :* PCR RA + pSB1A3 (no grow, but was messed up) | ||
| + | :* PCR DA + pSB1A3 (growth) | ||
| + | * Liquid cultures | ||
| + | :* 3 tubes total | ||
| + | :* RAGE, Se1GE, PCR DA in A3 (Amp broth) | ||
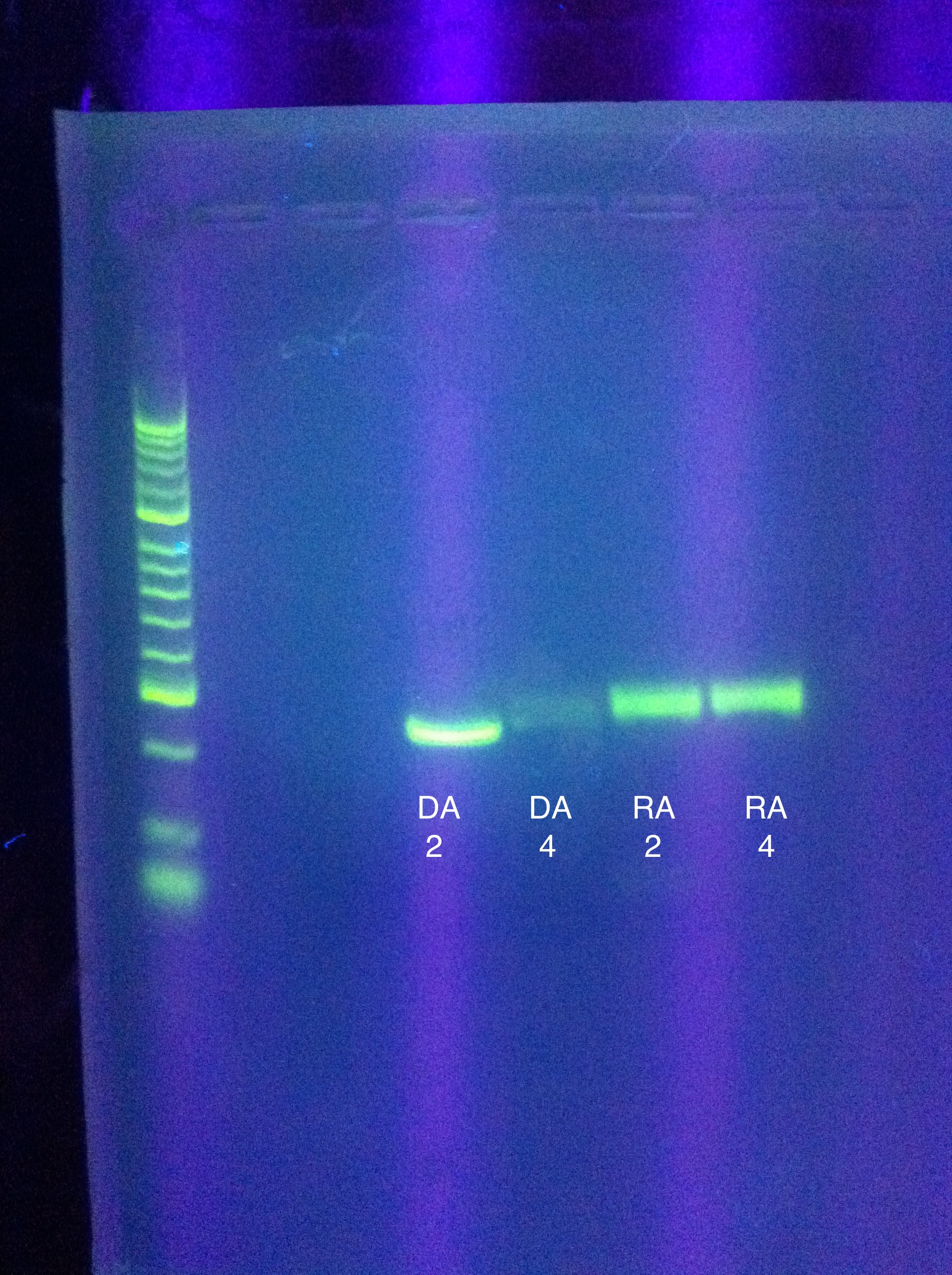
| + | * Gel of PCR results | ||
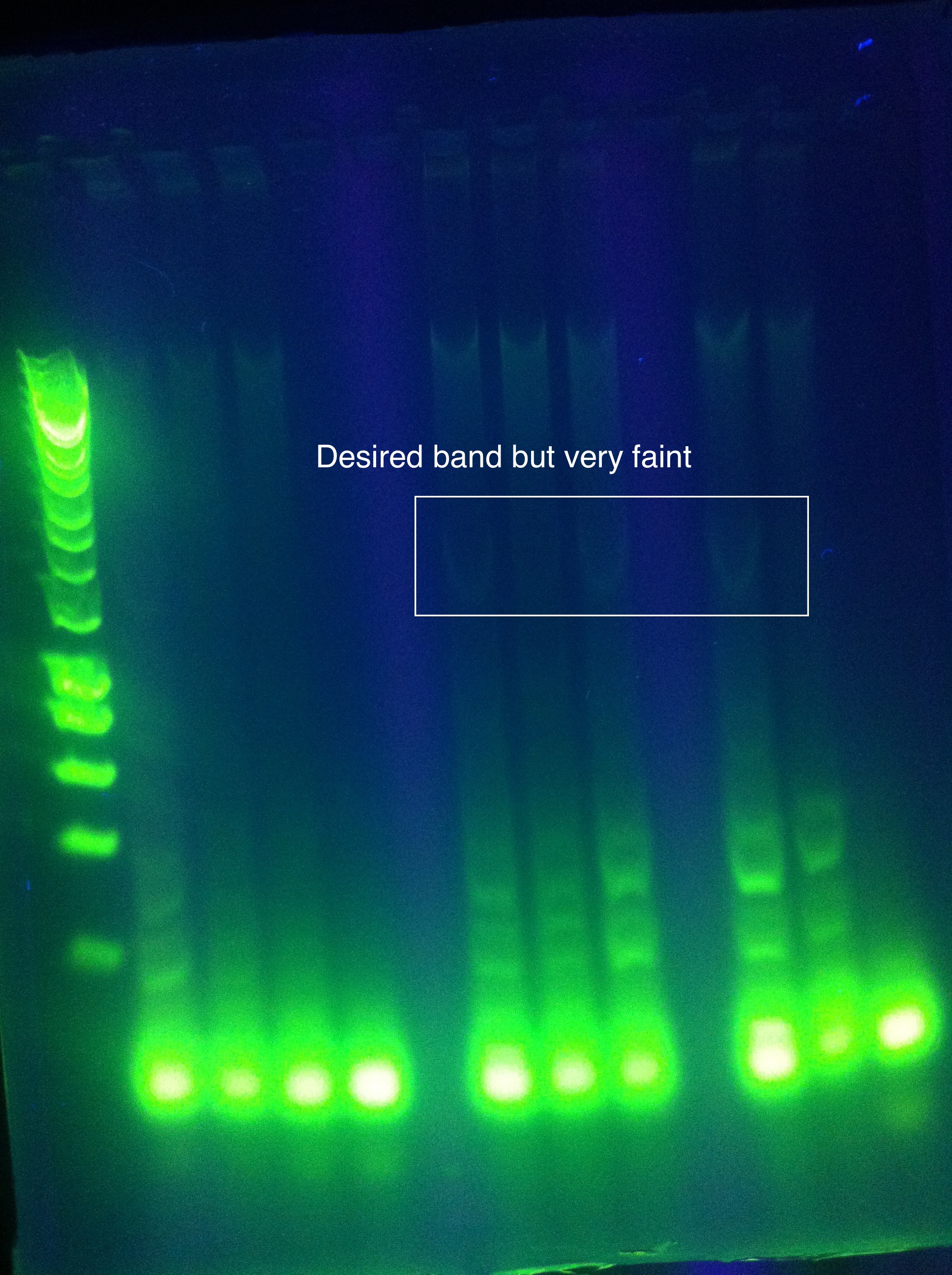
| + | <br> [[Image:ASU_7_25_gel_abcde.jpeg|400px]] | ||
| + | :* CasABCDE: not much visible, very faint and nonspecific | ||
| + | ::* Could be a result of changing the settings too much | ||
| + | ::* We expect our new primers today/tomorrow so we will likely wait for that | ||
| + | :* Cas3: lots of bands visible, looks as though there's a clear band at ~2600 | ||
| + | <br> [[Image:ASU_7_25_gel_cas3.jpeg|400px]] | ||
| + | ::* Gel extraction | ||
| + | * Miniprep of successful constitutive GFP generator | ||
| + | * To nanodrop | ||
| + | :* GFP generator | ||
| + | :* E0840 gel extract | ||
| + | * Restriction | ||
| + | :* Preparation of pSB1K3 backbone | ||
| + | ::* Protocol from Standard Parts Registry (E, P, DpnI) | ||
| + | :* RFP in pSB1A3 (EP) | ||
| + | * Ligation/Transformation | ||
| + | :* RFP + pSB1K3 | ||
| + | * Notes: | ||
| + | :* Still waiting on sequencing results to confirm that the things we have amplified are indeed RA, DA, and Seq1 ((though we won't be sure about DA because we didn't submit it yet)) | ||
| + | :* We can submit our 1x in A3 for sequencing tomorrow, after it grows up | ||
| + | == Tuesday, July 26 == | ||
| + | * Discussed USC CRISPER Conundrum | ||
| + | * Discussed alternate spacers to assemble in parallel | ||
| + | :* RFP | ||
| + | :* Nickase | ||
| + | :* Biofilm "Isr" | ||
| + | * Miniprep | ||
| + | :* 1xSeq1, RA, DA in psb1A3 | ||
| + | * Genome Prep | ||
| + | * PCR w/ new primers | ||
| + | :* CasABCDE - 68 degree annealing temp, 2 minute elongation | ||
| + | :* Cas3 - 62 degree annealing temp, 1:20 elongation | ||
| + | * Gel results: | ||
| + | :* ABCDE - mostly blank | ||
| + | :* Cas3 - broad nonspecific bands around desired area | ||
| + | * Nanodrop | ||
| + | :* 1x Seq1, RA, DA (400 ng/uL or so) | ||
| + | :* genome prep (77 ng/uL) | ||
| + | :* extracted cas3 (11 ng/uL) | ||
| + | :* GFP (4000 ng/uL - NEW FLAYPREP RECORD) | ||
| + | * Checked transformants | ||
| + | :* 1 of 2 gfp streak plates glows green, the other is partial | ||
| + | :* RFP plasmids are not red | ||
| + | ::* could be a result of improper AAP use, will try again | ||
| + | ::* grew up in a liquid culture (tried Kan and Amp separately) | ||
| + | :::* Did not grow in amp broth - no vector/vector ligation | ||
| + | * Restriction Digest | ||
| + | :* (EP) pSB1K3 backbone + AAP | ||
| + | :* (EP) RFP in pSB1A3 | ||
| + | * Ligation | ||
| + | :* 1:1 insert:vector ratio according to linearized plasmid backbone protocol | ||
| + | :* 3:1 insert to vector ratio according to OWW protocol | ||
| + | :* Both methods transformed/plated | ||
| + | * Overnight RE Digest | ||
| + | :* Seq1 in pSB1A3 (ES, XP) | ||
| + | :* DA in pSB1A3 (ES, XP) | ||
| + | :* RA in pSB1A3 (ES, XP) | ||
| + | :* (EP) pSB1K3 from "DA in pSB1K3", which is not really DA but is really the vector | ||
| + | ::* need to AAP this tomorrow | ||
| + | * Sequencing Results | ||
| + | :* Gel extracts of PCR amplified Seq1 and RA are good | ||
| + | * Sequencing Submission | ||
| + | * GFP forward | ||
| + | :* Cas3 F/R | ||
| + | :* Seq1, DA, RA 1x in psb1A3 (Forward/Reverse) | ||
| + | == Wednesday, July 27 == | ||
| + | * Ligate 2x in psb1k3 | ||
| + | :* Using overnight digests | ||
| + | ::* Seq1 in pSB1A3 (ES, XP) | ||
| + | ::* DA in pSB1A3 (ES, XP) | ||
| + | ::* RA in pSB1A3 (ES, XP) | ||
| + | ::* (EP) pSB1K3 from "DA in pSB1K3", which is not really DA | ||
| + | * Transformation of this | ||
| + | * Begin duet construction: | ||
| + | :* Transformed duet plasmid | ||
| + | * Streak plate RFP plates | ||
| + | * PCR: | ||
| + | :* CasABCDE with new and old primers and a temp gradient for both | ||
| + | :* New: 64->69, three rows of 3 tubes | ||
| + | :* Old: 63->66, three rows of 3 tubes | ||
| + | * Gel results: | ||
| + | <br> [[Image:ASU_727_IMG_3952.jpg|400px]] | ||
| + | :* New -- no good | ||
| + | :* Old -- mostly blank with some dimerization | ||
| + | * Made more LB + kan, amp, no antibiotic plates | ||
| + | * Talked about CMR things | ||
| + | :* We can buy P. furiosus DNA at 2 ug for $300 | ||
| + | == Thursday, July 28 == | ||
| + | * Plates from yesterday: | ||
| + | :* Negative control with amp: growing | ||
| + | :* RFP streak plates: correct | ||
| + | :* Duet vector: correct | ||
| + | :* 2x ra, da, seq1: did not grow | ||
| + | * Gel of restrictions from yesterday | ||
| + | :* 1st try: no success, ran off | ||
| + | :* 2nd try: no success, only backbones visible... | ||
| + | * PCR cleanup of DA PCRPCR to nanodrop, transform | ||
| + | * PCR: | ||
| + | :* CasABCDE (old primers) | ||
| + | ::* TDWN728B | ||
| + | ::* 71 to 68, -.1/cycle, 30 cycles | ||
| + | ::* 2:10 elongation | ||
| + | :* CasABCDE (new primers) | ||
| + | ::* TDWN728A | ||
| + | ::* 72 to 65, -.2/cycle, 35 cycles | ||
| + | ::* 2:10 elongation | ||
| + | * Gel results: | ||
| + | :* No good! ran these at night, all on one gel, and only one lane had anything at all and even then it was one large section of nonspecific amplification | ||
| + | * Meeting with grad students: | ||
| + | :* Homology between casA and human immune proteins (which ones?) | ||
| + | ::* Could be an evolutionary thing | ||
| + | :* Look into this could also be | ||
| + | ::* Conserved functional domains | ||
| + | == Friday, July 29 == | ||
| + | * Transformation results from yesterday | ||
| + | :* One colony of 1xDA was not pink, yay | ||
| + | * Remade liquid culture of 1xDA because morning miniprep had goopy crap | ||
| + | :* Will miniprep later | ||
| + | * RLT? | ||
| + | :* Will wait until we miniprep 1xDA | ||
| + | * Gibson Assembly? Do we have all materials? | ||
| + | :* No, we'll have to order a couple of things | ||
| + | :* Will plan on doing this and trying Gibson in parallel | ||
| + | * Made liquid culture of Duet plasmid | ||
| + | * Made more LB Kan media | ||
| + | * PCR | ||
| + | :* Retry original 7/19 run that was semi-successful | ||
| + | :* Another ABCDE | ||
| + | * Night Results | ||
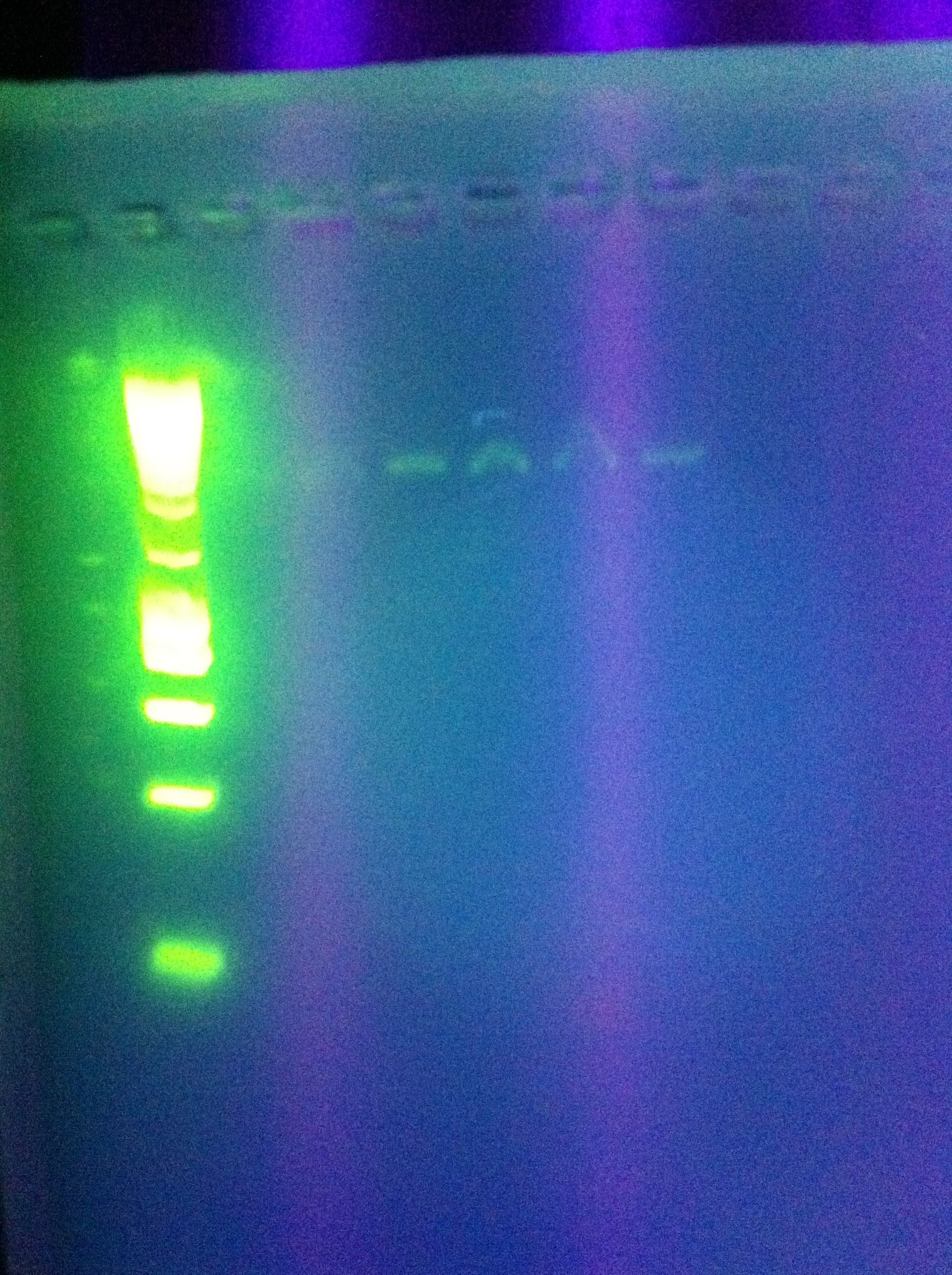
| + | :* Gels: No good. Nothing. "Dimer Central" | ||
| + | <br> [[Image:ASU_7_29_gel.jpg|400px]] | ||
| + | :* Miniprep of 1xDA | ||
| + | :* Made SOC mini-stocks | ||
| + | :* Glycerol stocks of something maybe | ||
| + | == Saturday, July 30 == | ||
| + | * PCR | ||
| + | :* "Touchup" PCR | ||
| + | * RLT | ||
| + | :* 2x DA, Seq1, RA (Ginkgo) | ||
| + | == Sunday, July 31 == | ||
| + | * Gel Results of "Touchup" | ||
| + | :* Worthless | ||
| + | * Check plates | ||
| + | :* No transformants but perhaps a few small ones | ||
| + | * Redo transformation | ||
| + | * Redigest RFP pSB1K3 for another RLT | ||
| + | }} | ||
Latest revision as of 02:54, 29 September 2011
|
|
Friday, July 1
Saturday, July 2
Tuesday, July 5
Wednesday, July 6
Thursday, July 7
Friday, July 8
Sunday, July 10
Monday, July 11
Tuesday, July 12
Wednesday, July 13
Thursday, July 14
Friday, July 15
Saturday, July 16
Sunday, July 17
Monday, July 18
Tuesday, July 19
Wednesday, July 20
Thursday, July 21
Friday, July 22
Saturday, July 23
Sunday, July 24
Monday, July 25
Tuesday, July 26
Wednesday, July 27
Thursday, July 28
Friday, July 29
Saturday, July 30
Sunday, July 31
|
 "
"