Team:Arizona State/Project/Software
From 2011.igem.org
(Difference between revisions)
Ethan ward (Talk | contribs) |
Ethan ward (Talk | contribs) |
||
| Line 9: | Line 9: | ||
'''Downloads (coming soon)''' | '''Downloads (coming soon)''' | ||
---- | ---- | ||
| - | * Python source: | + | * Python source: [http://code.google.com/p/crispr-studio/ Google code development page] |
:* Dependencies: BioPython, NumPy, WxPython, BLAST+ | :* Dependencies: BioPython, NumPy, WxPython, BLAST+ | ||
* Executable: | * Executable: | ||
Revision as of 06:58, 27 September 2011
|
|
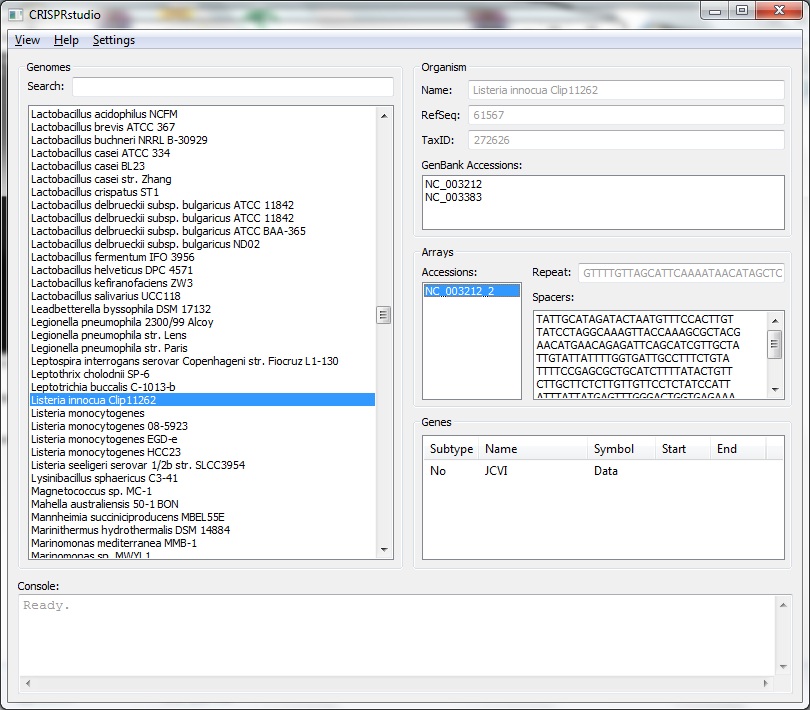
We have developed a tool to assist in the development of synthetic CRISPR systems.
Downloads (coming soon)
Sources
|
 "
"