Team:Arizona State/Project/Software
From 2011.igem.org
(Difference between revisions)
Ethan ward (Talk | contribs) |
Ethan ward (Talk | contribs) |
||
| Line 1: | Line 1: | ||
{{:Team:Arizona State/Templates/main|title=CRISPR Studio|content= | {{:Team:Arizona State/Templates/main|title=CRISPR Studio|content= | ||
| | ||
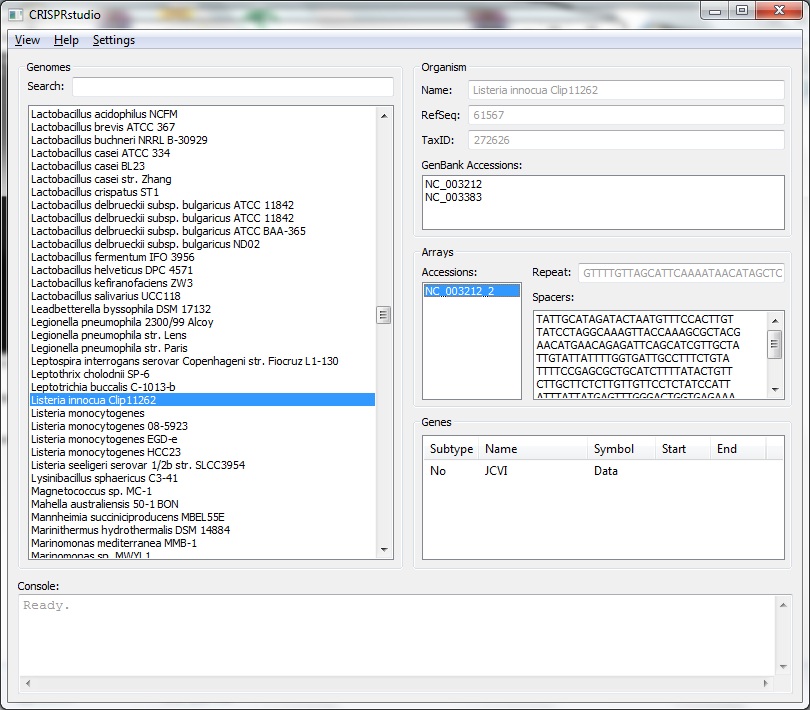
| - | [[File:ASU crisprstudio ss.jpg|thumb]] | + | [[File:ASU crisprstudio ss.jpg|thumb|CRISPR information panel of the CRISPRstudio]] |
We have developed a tool to assist in the development of synthetic CRISPR systems. | We have developed a tool to assist in the development of synthetic CRISPR systems. | ||
* Pick spacers from a source sequence, based on homology with the target genome, hairpinning potential, restriction sites, and known PAMs. | * Pick spacers from a source sequence, based on homology with the target genome, hairpinning potential, restriction sites, and known PAMs. | ||
| Line 10: | Line 10: | ||
---- | ---- | ||
* Python source: | * Python source: | ||
| - | :* Dependencies: BioPython, NumPy, WxPython | + | :* Dependencies: BioPython, NumPy, WxPython, BLAST+ |
* Executable: | * Executable: | ||
| + | |||
'''Sources''' | '''Sources''' | ||
---- | ---- | ||
| Line 17: | Line 18: | ||
* [http://cmr.jcvi.org/tigr-scripts/CMR/CmrHomePage.cgi JCVI CMR] | * [http://cmr.jcvi.org/tigr-scripts/CMR/CmrHomePage.cgi JCVI CMR] | ||
* [http://crispi.genouest.org/ CRISPI] | * [http://crispi.genouest.org/ CRISPI] | ||
| - | + | * NCBI | |
}} | }} | ||
Revision as of 04:09, 27 September 2011
|
|
We have developed a tool to assist in the development of synthetic CRISPR systems.
Downloads (coming soon)
Sources
|
 "
"