Team:Harvard/Results
From 2011.igem.org
(Difference between revisions)
| Line 4: | Line 4: | ||
</div> | </div> | ||
<div class="whitebox"> | <div class="whitebox"> | ||
| + | __NOTOC__ | ||
=Bioinformatics= | =Bioinformatics= | ||
==55,000 Possible Zinc Fingers== | ==55,000 Possible Zinc Fingers== | ||
| Line 16: | Line 17: | ||
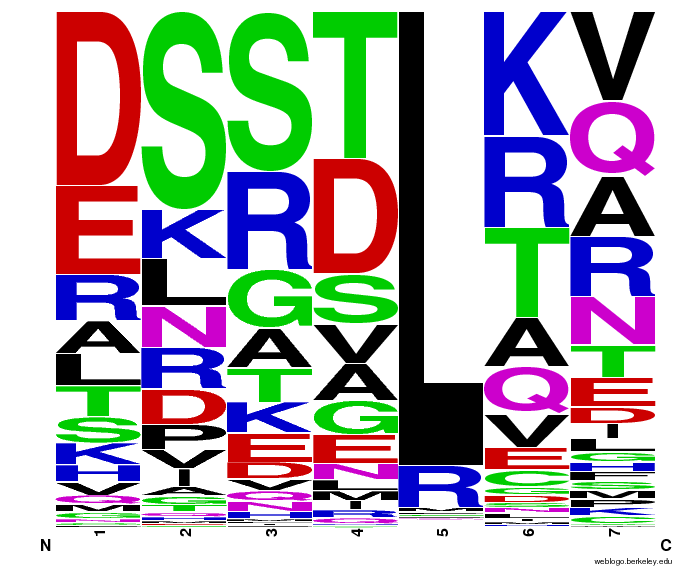
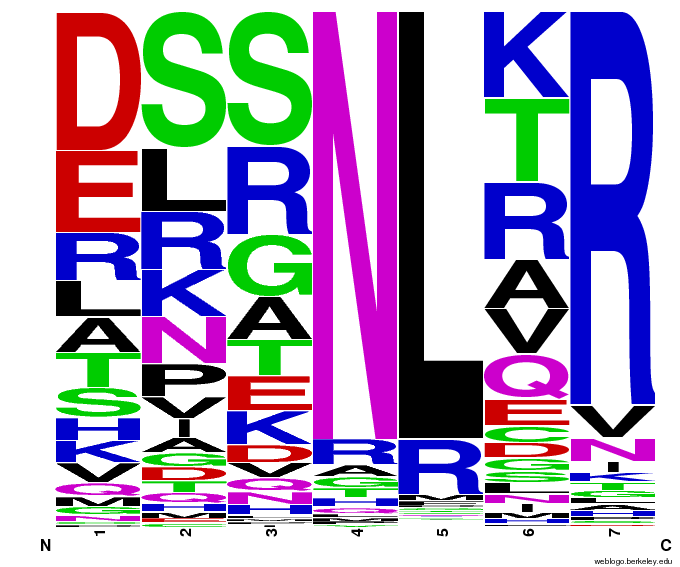
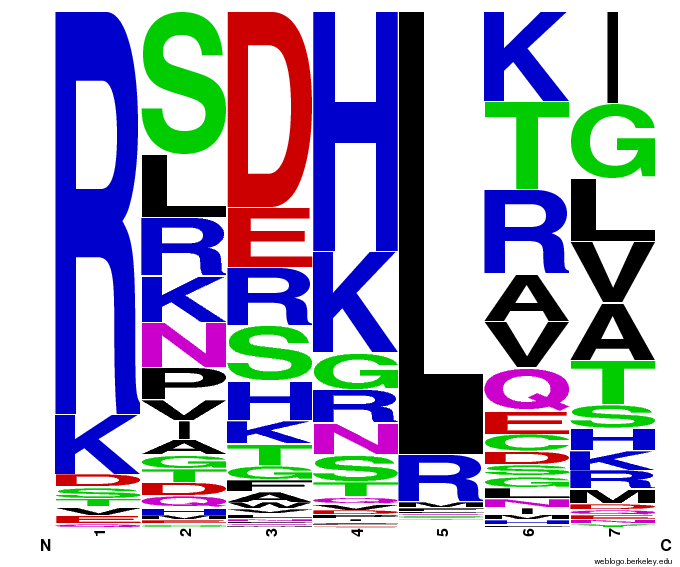
| [[File:HARVCTG.png|thumb|left|CTG]]|| [[File:HARVGAC.png|thumb|left|GAC]]}|| [[File:HARVTGG.png|thumb|left|TGG]] | | [[File:HARVCTG.png|thumb|left|CTG]]|| [[File:HARVGAC.png|thumb|left|GAC]]}|| [[File:HARVTGG.png|thumb|left|TGG]] | ||
|} | |} | ||
| - | |||
</div> | </div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
Revision as of 15:38, 25 September 2011
Overview | MAGE | Lambda Red| Chip-Based Library | One-Hybrid Selection | Zinc Finger Binders | Biobricks
Bioinformatics
55,000 Possible Zinc Fingers
We made 55,000 sequences, distributed evenly among 6 DNA target triplets. That's 9150 per target.
Because our program's output changes dramatically based on the input triplet, no two sets of sequences are the same:
| } |
 "
"