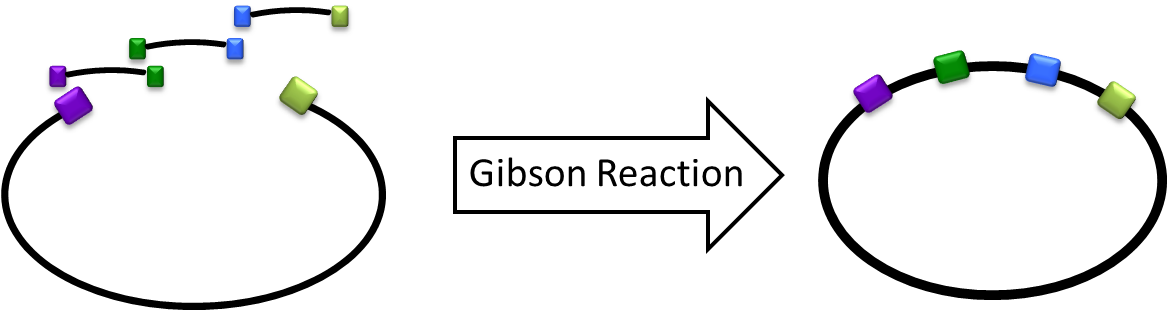Team:Washington/Magnetosomes/GibsonVectors
From 2011.igem.org
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
| - | + | ==Gibson Vector Toolkit== | |
| - | ==About Gibson Cloning/Assembly== | + | ===About Gibson Cloning/Assembly=== |
| - | Gibson Cloning/Assembly is a cloning method that allows multiple inserts in one time isothermal reaction for assembling overlapping DNA fragments. During the cloning process, this does not only saves a lot of time and efforts, but also allows more complexity when building the circuit. | + | Gibson Cloning/Assembly is a cloning method that allows multiple inserts in one time isothermal reaction for assembling overlapping DNA fragments. During the cloning process, this does not only saves a lot of time and efforts, but also allows more complexity when building the circuit. <br/> [[File:Igem2011_GibsonReacion.png|center|400px]] |
| - | ==What happened last year?== | + | ===What happened last year?=== |
The Gibson Cloning method is definitely not a new terminology to the iGEM community. This cloning method was published in Nature protocols 2009 ( Please see Enzymatic assembly of DNA molecules up to several hundred kilobases, Gibson et al. (2009).) and it has gained a lot of attention from the iGEM community since then. | The Gibson Cloning method is definitely not a new terminology to the iGEM community. This cloning method was published in Nature protocols 2009 ( Please see Enzymatic assembly of DNA molecules up to several hundred kilobases, Gibson et al. (2009).) and it has gained a lot of attention from the iGEM community since then. | ||
| Line 18: | Line 18: | ||
*E=ECoRI,S1=Spacer 1, Bg=Bgl S2=Spacer2, P=PstI | *E=ECoRI,S1=Spacer 1, Bg=Bgl S2=Spacer2, P=PstI | ||
| - | |||
| - | Seeing that this is such a great method to do cloning...we | + | === What about this year? === |
| + | |||
| + | Seeing that this is such a great method to do cloning...we continued with the investigation and made the ''Gibson Assembly Toolkit''! | ||
| + | |||
| + | We call our new vectors plamid Gisbon Assembly (pGA) vectors. And we were able to show that the cloning efficiency of pGA vector is better than the pSB vector. | ||
| + | |||
| + | (Stay tuned for our results) | ||
| + | |||
| + | |||
| + | We submitted 5 pGA vectors of different copy numbers and antibiotics resistance to the Registry. (All of them have pLac GFP) | ||
| + | |||
| + | pGA1A3, pGA1C3 are high copy plasmid backbone, which are good for 1 or 2 gene inserts. | ||
| + | pGA 3K3, pGA 4C5 and pGA 4A5 are low copy plasmid backbone, which are good for more gene fragment inserts. | ||
| + | |||
| + | |||
| - | |||
a diagram | a diagram | ||
a table showing the antibiotics, copy number | a table showing the antibiotics, copy number | ||
| - | == Next level of iGEM project: more complex circuit.== | + | === Next level of iGEM project: more complex circuit.=== |
And we decided to look at magnetosome | And we decided to look at magnetosome | ||
Revision as of 22:11, 22 September 2011
Gibson Vector Toolkit
About Gibson Cloning/Assembly
Gibson Cloning/Assembly is a cloning method that allows multiple inserts in one time isothermal reaction for assembling overlapping DNA fragments. During the cloning process, this does not only saves a lot of time and efforts, but also allows more complexity when building the circuit.What happened last year?
The Gibson Cloning method is definitely not a new terminology to the iGEM community. This cloning method was published in Nature protocols 2009 ( Please see Enzymatic assembly of DNA molecules up to several hundred kilobases, Gibson et al. (2009).) and it has gained a lot of attention from the iGEM community since then.
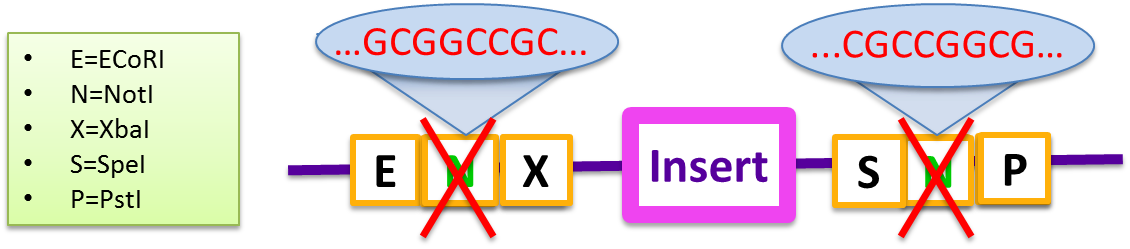
In 2010, the Cambridge iGEM team proposed the proposed a BioBrick standard for Gibson Assembly (RFC 57) which enables simultaneous assembly of multiple fragments with no scar sequences. We found that this Biobrick Standard was incapable of giving high yields even in two-fragments assembly. The primary problem is the self-complementarity of the two NotI sequences embedded in the BioBrick prefix and suffix which prevents the insert from being incorporated into the vector.
- E=ECoRI, N=NotI, X=XohI, S=SpeI, P=PstI
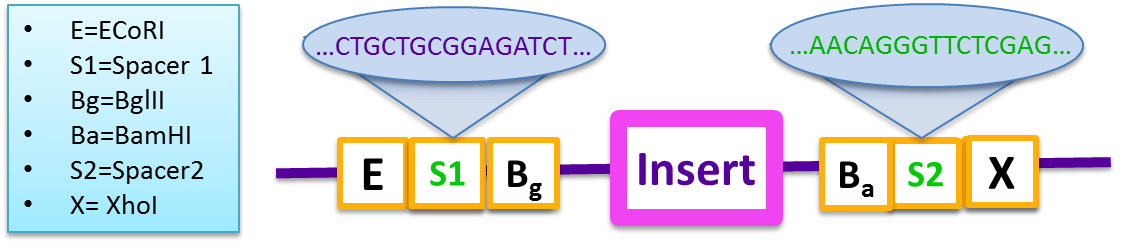
Therefore, to combat the problem, a “gibson reaction compatible” prefix and suffix were designed based on BglBrick standard to increase the cloning efficiency.
- E=ECoRI,S1=Spacer 1, Bg=Bgl S2=Spacer2, P=PstI
What about this year?
Seeing that this is such a great method to do cloning...we continued with the investigation and made the Gibson Assembly Toolkit!
We call our new vectors plamid Gisbon Assembly (pGA) vectors. And we were able to show that the cloning efficiency of pGA vector is better than the pSB vector.
(Stay tuned for our results)
We submitted 5 pGA vectors of different copy numbers and antibiotics resistance to the Registry. (All of them have pLac GFP)
pGA1A3, pGA1C3 are high copy plasmid backbone, which are good for 1 or 2 gene inserts. pGA 3K3, pGA 4C5 and pGA 4A5 are low copy plasmid backbone, which are good for more gene fragment inserts.
a diagram a table showing the antibiotics, copy number
Next level of iGEM project: more complex circuit.
And we decided to look at magnetosome
 "
"