Team:Paris Bettencourt/Experiments/tRNA diffusion
From 2011.igem.org
(Difference between revisions)
(→tRNA diffusion) |
|||
| Line 1: | Line 1: | ||
{{:Team:Paris_Bettencourt/tpl_test}} | {{:Team:Paris_Bettencourt/tpl_test}} | ||
| + | |||
| + | ==Results== | ||
| + | |||
| + | We managed to Characterize the tRNA biobrick by taking pictures of two constructs. | ||
| + | ===GFP amber=== | ||
| + | One of our strains contains only GFP amber, without the tTRNA corresponding. Cells do not glow. this result is a reliable negative control. | ||
| + | |||
| + | ===GFP amber+tRNA amber=== | ||
| + | The second strain contains the full construct and the production of tRNA is ruled by PHyperspank which is activated by IPTG. | ||
| + | In order to make a negative control we plated them without iptg. Cells glows a bit. Moreover some of the cells strongly leak. | ||
| + | |||
| + | ===GFP amber + tRNA amber + IPTG=== | ||
| + | The last result is given by testing the full construct with IPTG induced cells. Each cell glows in an almost equivalent way. The luminance is significant. | ||
| + | |||
| + | |||
| + | ==Characterization of tRNA biobrick== | ||
| + | |||
| + | |||
| + | |||
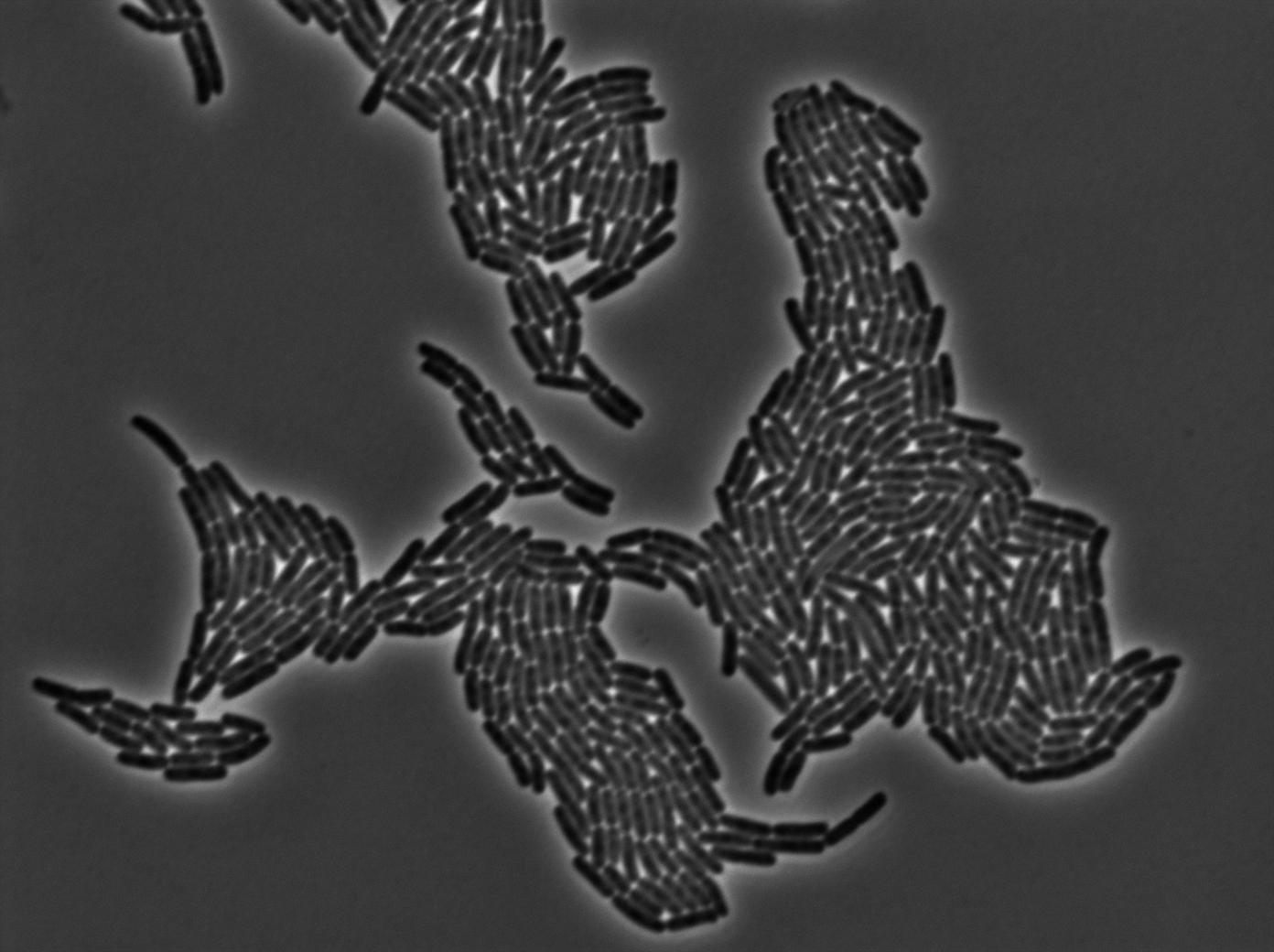
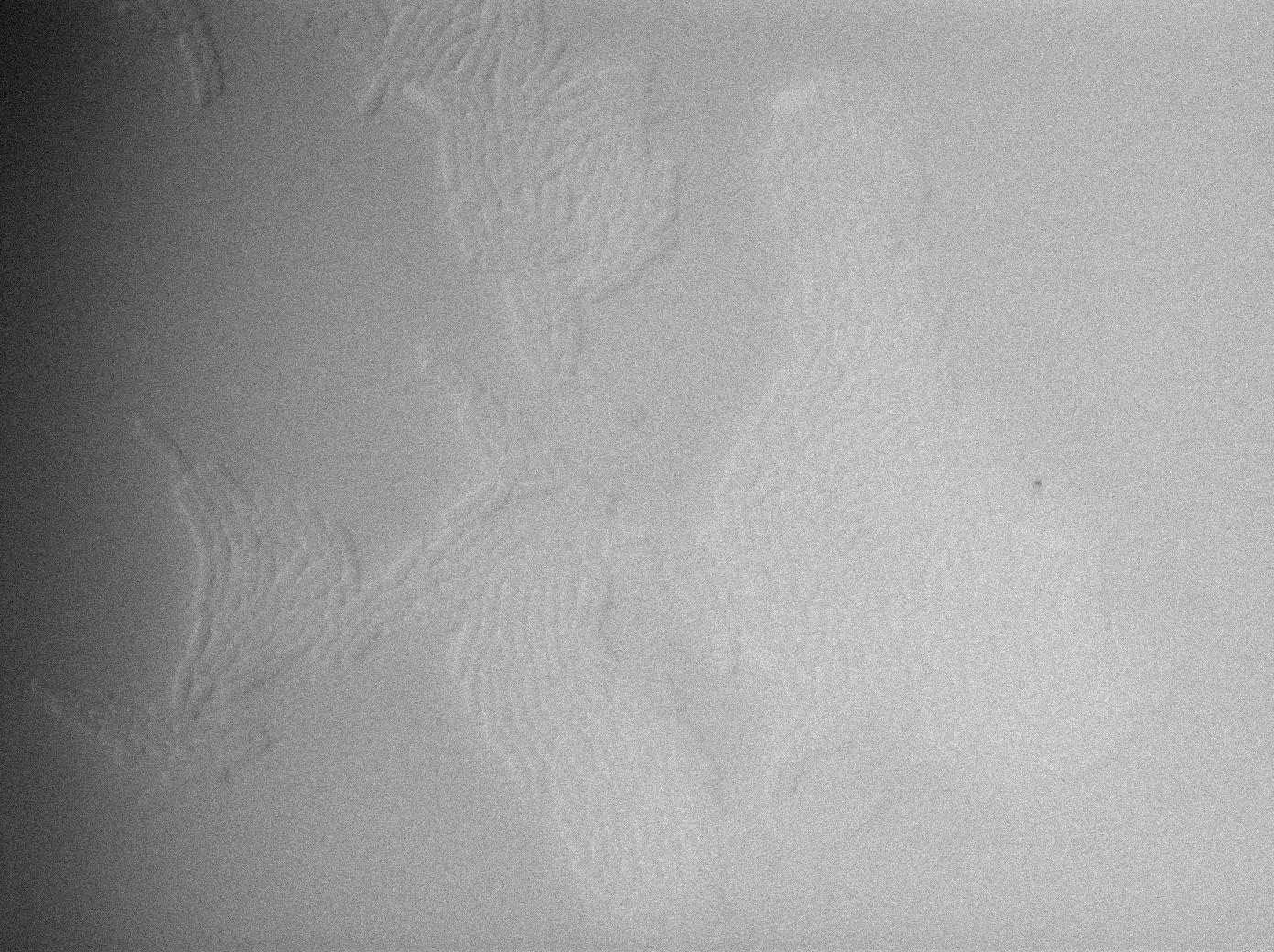
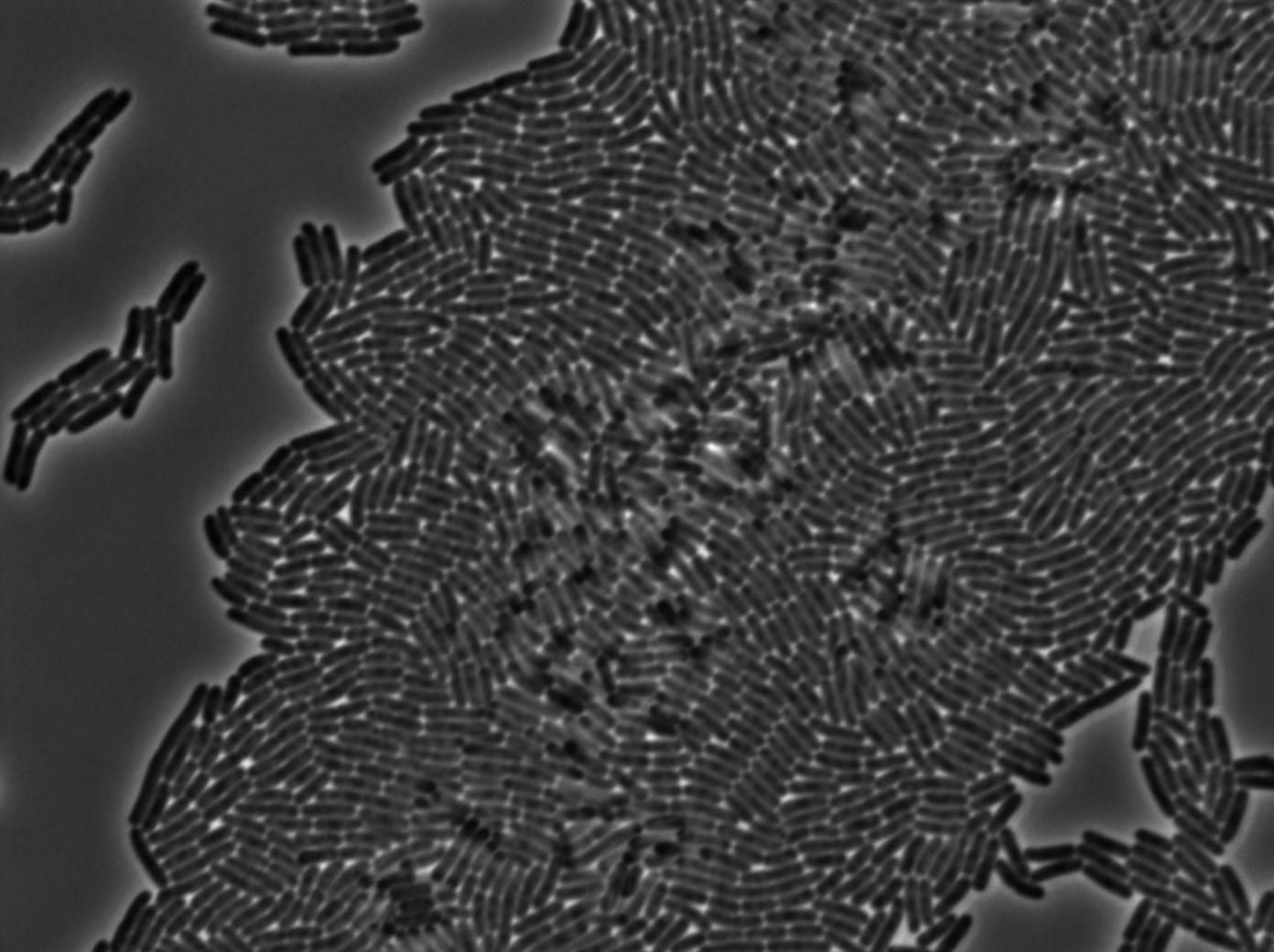
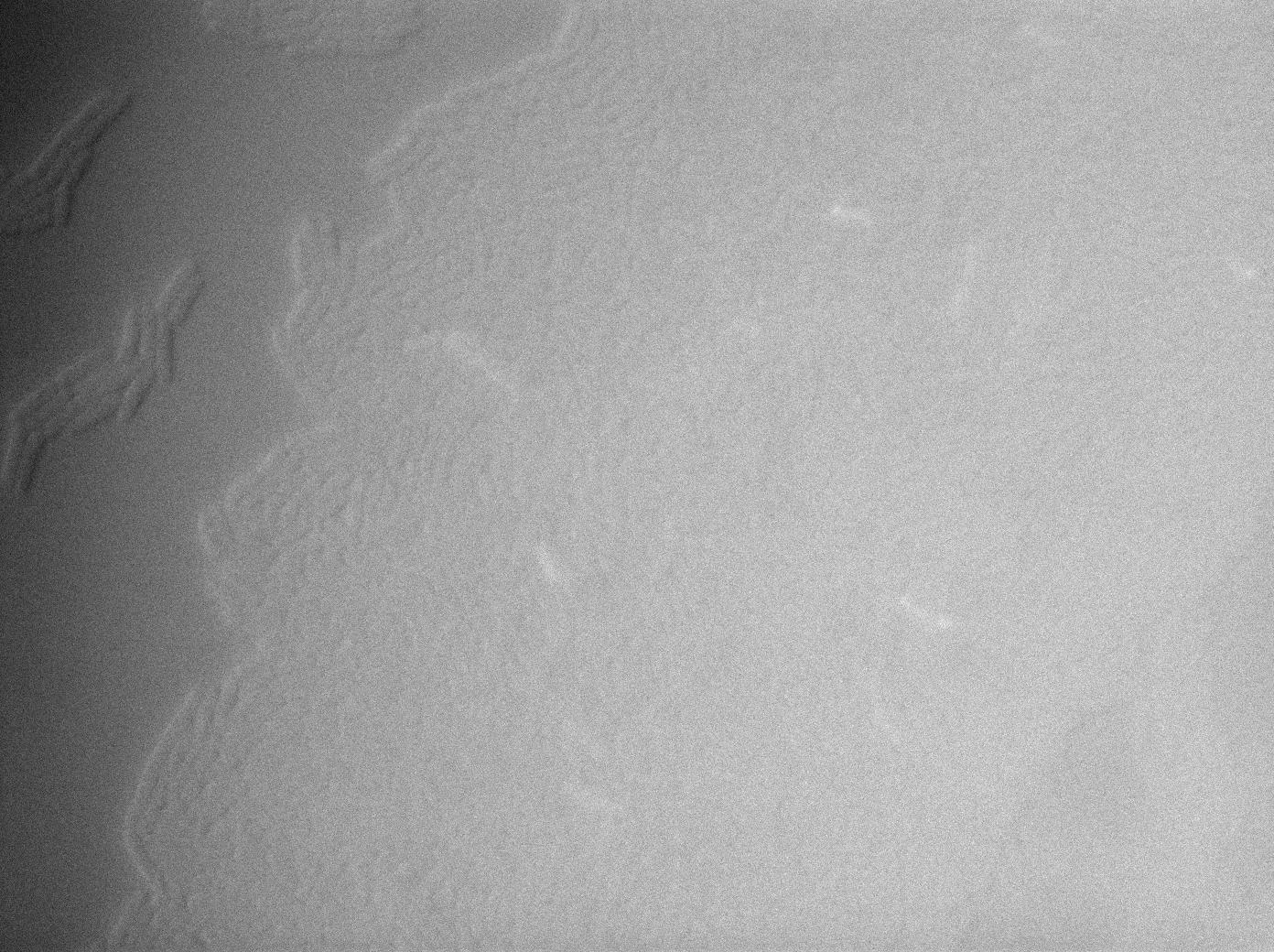
| + | {| border="1" class="wikitable" style="text-align: center;" | ||
| + | |+GFP amber / E-coli at 37°C | ||
| + | |- | ||
| + | |[[File:Trnaminus gfp amber trans 1.jpg|450px|thumb|center|E-Coli at 37°C (trans image)]] | ||
| + | |[[File:Trnaminus gfp amber fluo 1.jpg|450px|thumb|center|E-Coli at 37°C (gfp image)]] | ||
| + | |- | ||
| + | |[[File:Trnaminus gfp amber trans 2.jpg|450px|thumb|center|E-Coli at 37°C (trans image)]] | ||
| + | |[[File:Trnaminus gfp amber fluo 2.jpg|450px|thumb|center|E-Coli at 37°C (gfp image)]] | ||
| + | |} | ||
<html> | <html> | ||
| - | |||
<!-- PAGE FOOTER -- ITEMS FROM COLUMN ! HAVE BEEN MOVED HERE -- RDR --> | <!-- PAGE FOOTER -- ITEMS FROM COLUMN ! HAVE BEEN MOVED HERE -- RDR --> | ||
Revision as of 21:29, 21 September 2011

Contents |
Results
We managed to Characterize the tRNA biobrick by taking pictures of two constructs.
GFP amber
One of our strains contains only GFP amber, without the tTRNA corresponding. Cells do not glow. this result is a reliable negative control.
GFP amber+tRNA amber
The second strain contains the full construct and the production of tRNA is ruled by PHyperspank which is activated by IPTG. In order to make a negative control we plated them without iptg. Cells glows a bit. Moreover some of the cells strongly leak.
GFP amber + tRNA amber + IPTG
The last result is given by testing the full construct with IPTG induced cells. Each cell glows in an almost equivalent way. The luminance is significant.
Characterization of tRNA biobrick
 "
"