Team:DTU-Denmark/Background sRNA
From 2011.igem.org
m (moved Team:DTU-Denmark/sRNA to Team:DTU-Denmark/Background sRNA) |
|||
| Line 1: | Line 1: | ||
| - | {{:Team:DTU-Denmark/Templates/Standard_page_begin| | + | {{:Team:DTU-Denmark/Templates/Standard_page_begin|Background}} |
==What is sRNA== | ==What is sRNA== | ||
Revision as of 18:28, 21 September 2011
Background
Contents |
What is sRNA
In our project we dig into the fascinating world of sRNA regulation with the aim of engeneering a so-called trap sRNA regulating module and hereby creating a universal tool for a whole new way of regulating gene expression in synthetic biology. To fully understand our project, at basic knowledge of sRNAs, the way they regulate gene expression, the players involved (the RNA chaperone Hfq) and the unique features of the regulation is required. So continue reading if these words are new to you.
An recurring theme for sRNA regulation is the diversity of its mechanism; some sRNAs are repressors other activators, some act catalytically others non-catalytically, some require Hfq others not and so on. All in all this illustrates the extensive variety of sRNAs and how they have been tuned to operate in a range of regulatory niches in the cell during the process of evolution.
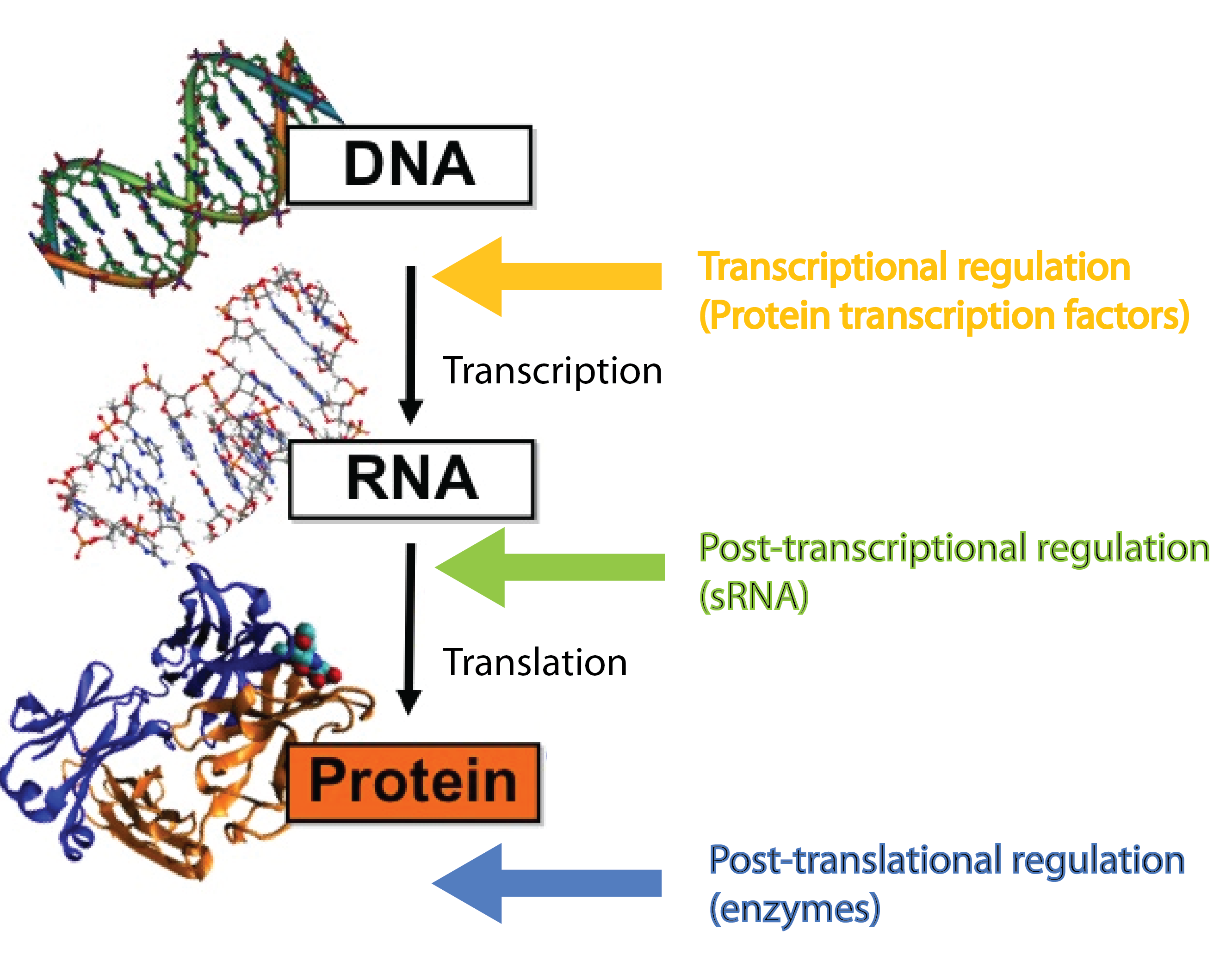
Many different kinds of RNA molecules are present in the cell; some of these are never translated into proteins. This group is collectively called non-coding RNAs, and includes, in addition to small RNA (sRNA), also rRNA and tRNA as well as more exotic RNA classes. Bacterial sRNAs are small RNA molecules (normally 50-200 nucleotides) which are extremely interesting as they have been demonstrated to play central regulatory roles in prokaryotes. There are two groups of sRNAs; the first group regulates gene expression by binding to proteins (e.g. transcription factors) and inhibiting their function, the second group regulates gene expression by binding to mRNAs. In our project we are focusing solely on the latter group, which regulate gene expression at the post-transcriptional level[2][4]. Whenever we mention sRNA we refer to those whit mRNA targets. Regulatory RNAs are not restricted to prokaryotes. Eukaryotes (including mammals) use micro RNA (miRNA) in post-transcriptional regulation, which is a functional analog to sRNA[7].
sRNAs act at the post-transcriptional level, a level complementary to the level at which protein regulators operate. Protein transcription factors operate before sRNAs at the transcriptional level whereas enzymes such as kinases and phosphatases act after sRNAs at the post-translational level[7]. Combining different layers of regulation enables interesting regulatory outcomes, eg. increase the number of genes regulated in response to a given signal or extremely tight repression of genes - sRNA can silence leaky promoters[7]. Transcription factors typically bind the target gene within ~100 nucleotides of the -10 and -35 region which set an upper limit for the number of transcription factors that can regulate a given gene. By targeting a complete different part of the gene, sRNAs can increase the number of signals a given gene can sense.
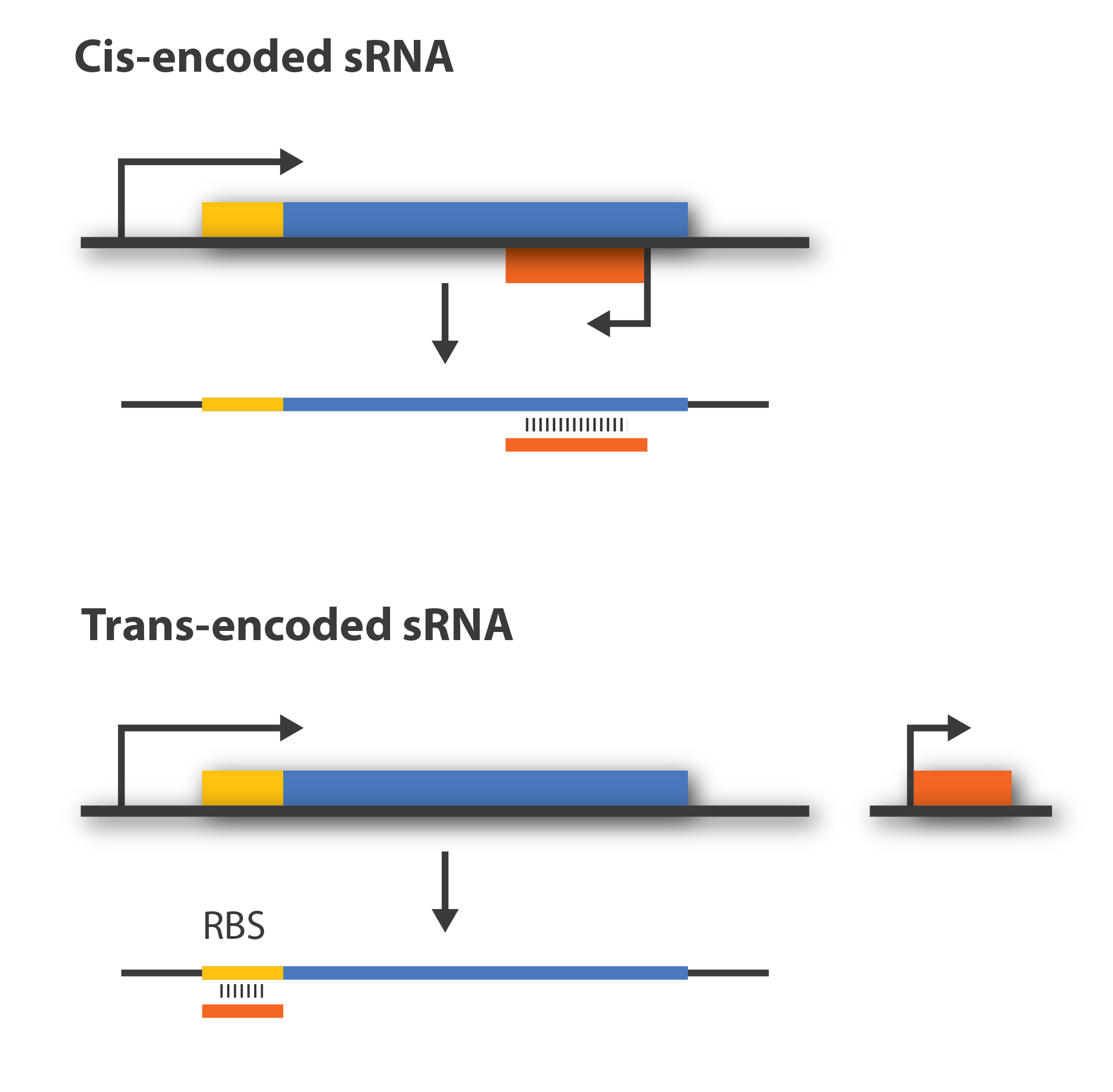
Cis-acting versus trans-acting sRNA
Regulatory sRNAs are divided into different sub-groups depending on their genomic locations with respect to its mRNA target. Cis-encoded or antisense sRNAs are encoded just opposite the one gene they regulate whereas trans-encoded sRNAs are located somewhere else in the genome and can aim at multiple number of target mRNA. Antisense RNA share extensive sequence complementarity with its target mRNA and hence do not require the RNA chaperone, Hfq, to stabilize the complex. This is in contrast to trans-encoded sRNAs which often only posses a complementary base-pairing region of 6-20 nucleotides and often require Hfq[5]. We are focusing on trans-encoded sRNA in the project, so whenever we write sRNA we implicit mean trans-encoded sRNA.
sRNA in Nature
Trans-encoded sRNAs are widespread in nature and an increasing number of sRNAs have been found to regulate critical pathways. In prokaryotes, sRNAs are predominantly involved in response to environmental stimuli or stress situations, eg. nutrient starvation, quorum sensing, membrane stress, oxidative stress, and SOS response to DNA damage[2][4]. Examples of well-known sRNAs include: the E.coli RyhB which downregulates non-essential iron-sulfur containing enzymes when iron is limited and the Vibrio Qrr which represses quorum sensing when the cell density is low[7].
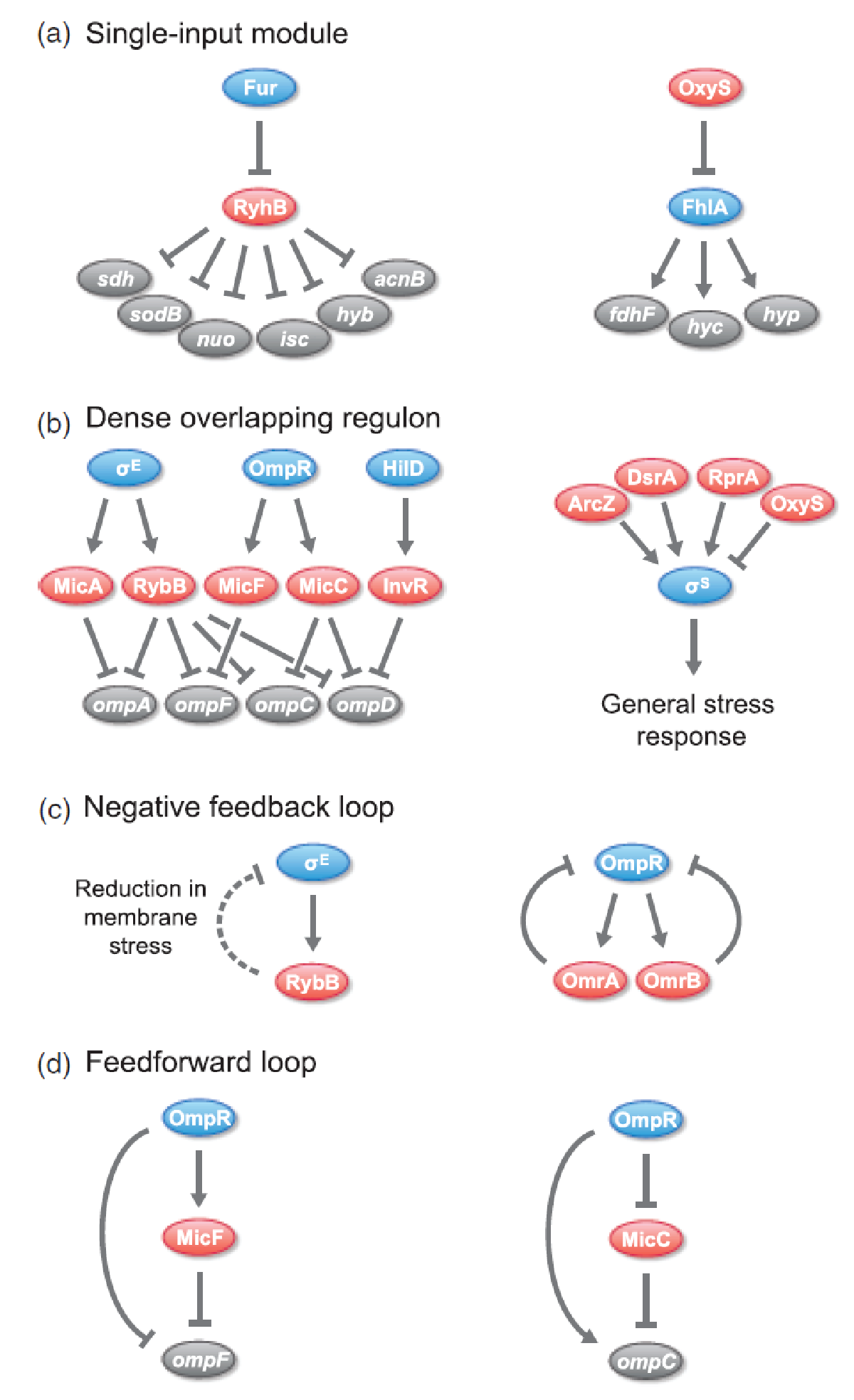
sRNAs have been found to be incorporated in various regulatory circuits which is similar to network motifs found in transcriptional regulatory networks, eg. single-input module (SIM), dense overlapping regulon (DOR), positive feedback loop, negative feedback loop, and feedforward loop (for those geeky guys into gene regulation)[2].
How do (trans) sRNAs Work as Regulators
The key-word for sRNA mediated regulation is base-pairing. A part of the sRNA shows limited complementary to the mRNA of the gene it regulates and the sRNA can thus base-pair with the mRNA. The region of base-pairing is typically only 6-20 nucleotides, and often it is only a subset of these nucleotides which seems to be critical for regulation[5][7]. This (often imperfect) base-pairing between the sRNA and target mRNA leads to changes in mRNA translation or stability - or both, and thereby influences the target gene expression[2]. sRNAs can be both activators and repressors for gene expression depending on what part of the mRNA molecule they base-pair with.
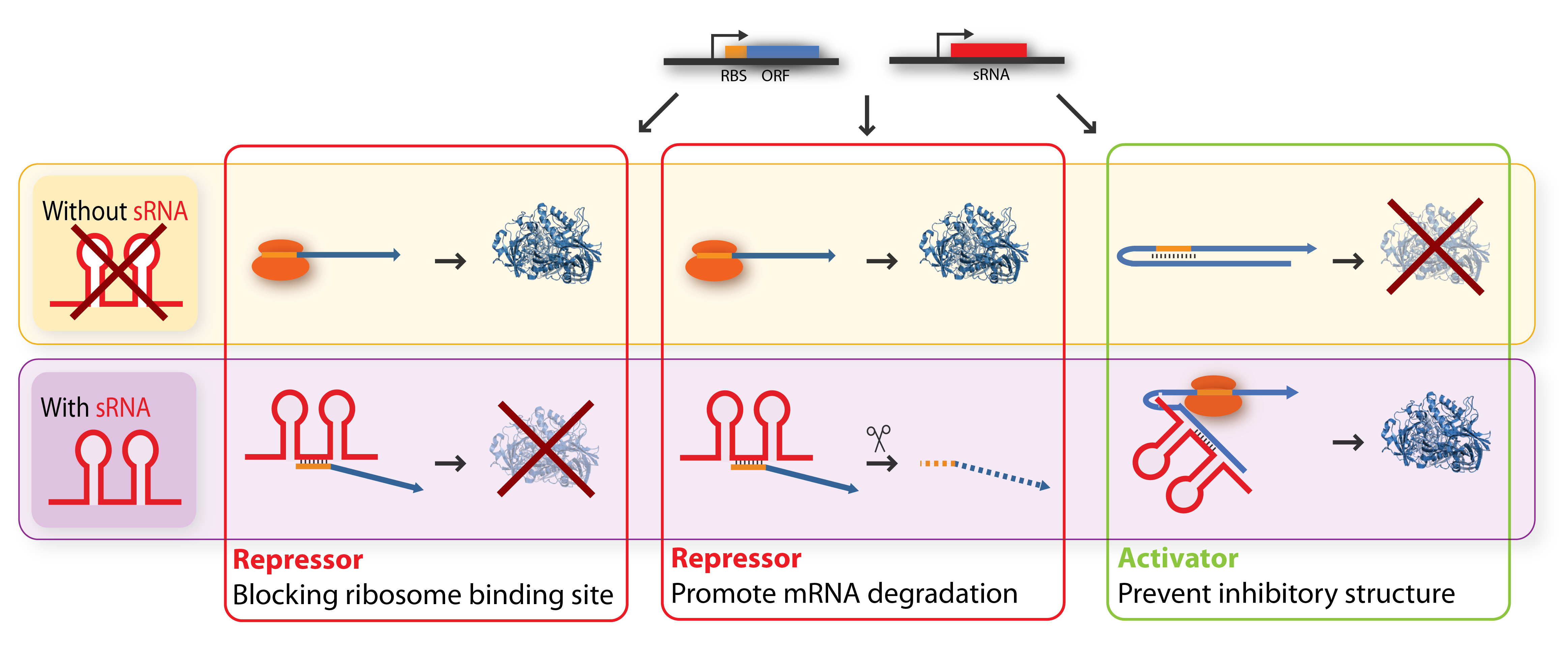
Repressor sRNAs act negatively by binding to the 5’ UTR often near the ribosome binding site. The binding inhibits translation by impeding ribosome binding and/or target the mRNA for degradation by RNases (often RNase E). Note that the degradation entails irreversible regulation[7]. Some sRNAs act stoichiometrically meaning that they are co-degradated with the mRNA whereas others act catalytically and are not degraded in the reaction.
Activator sRNAs act positively through an anti-antisense mechanism where sRNA base-pairing with the target mRNA disrupt an inhibitory secondary structure sequestering the ribosome-binding site. As a result, the ribosome-binding site is liberated and free to bind ribosomes[7].
The Role of Hfq
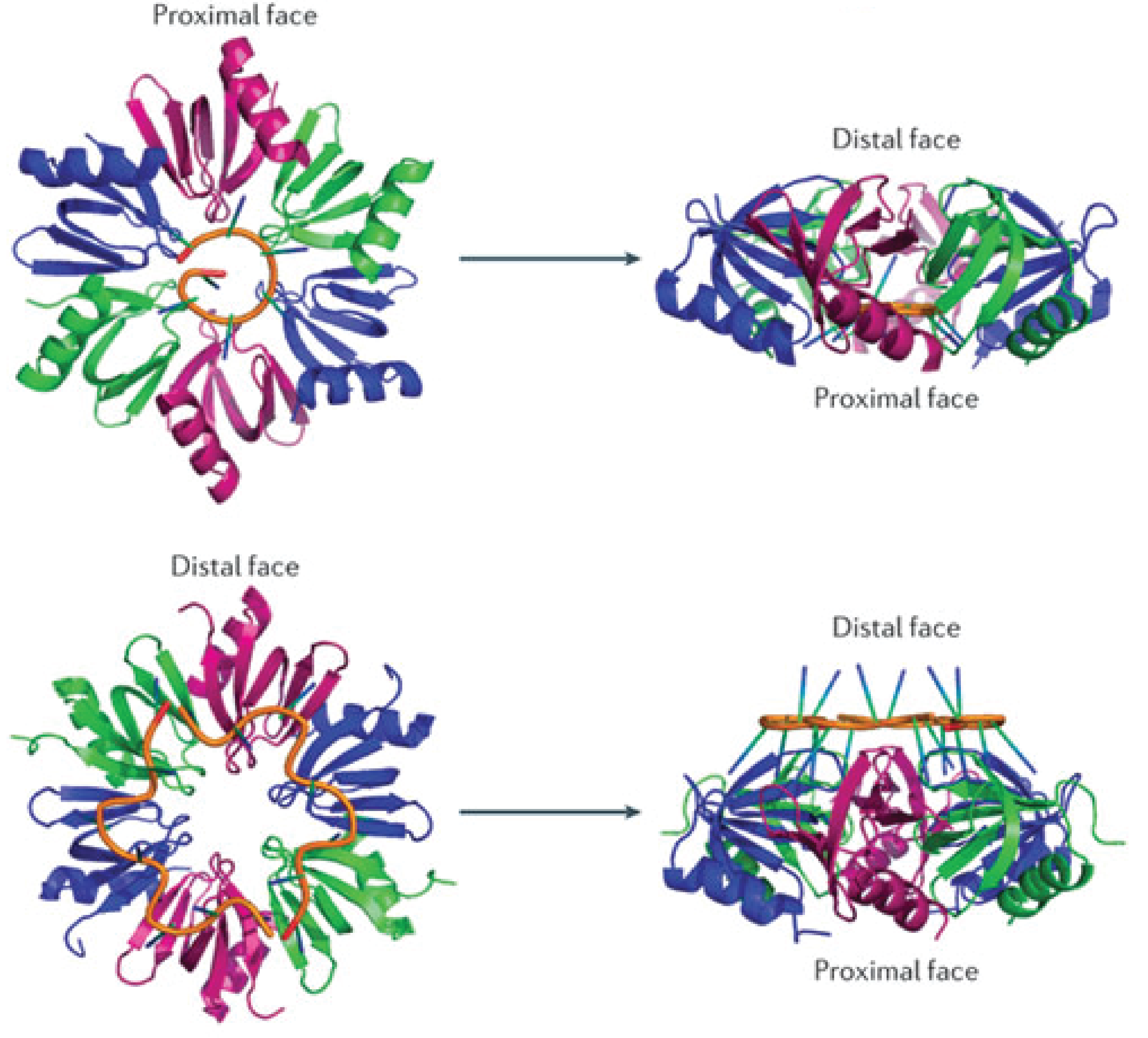
Hfq is an RNA-binding protein encoded in approximately half of the (in 2002) sequenced bacterial genomes and is one of the most abundant proteins in E.coli[3]. Hfq consists of six identical subunits arranged in a ringshape structure[3] with two RNA-binding faces and a long unstructured C-terminal. The sequence motif for RNA binding to the distal face is 5'-AAYAAYAA-3' (where Y is a pyrimidine, U or C), whereas the proximal face seems to favor U-rich sequences[6].
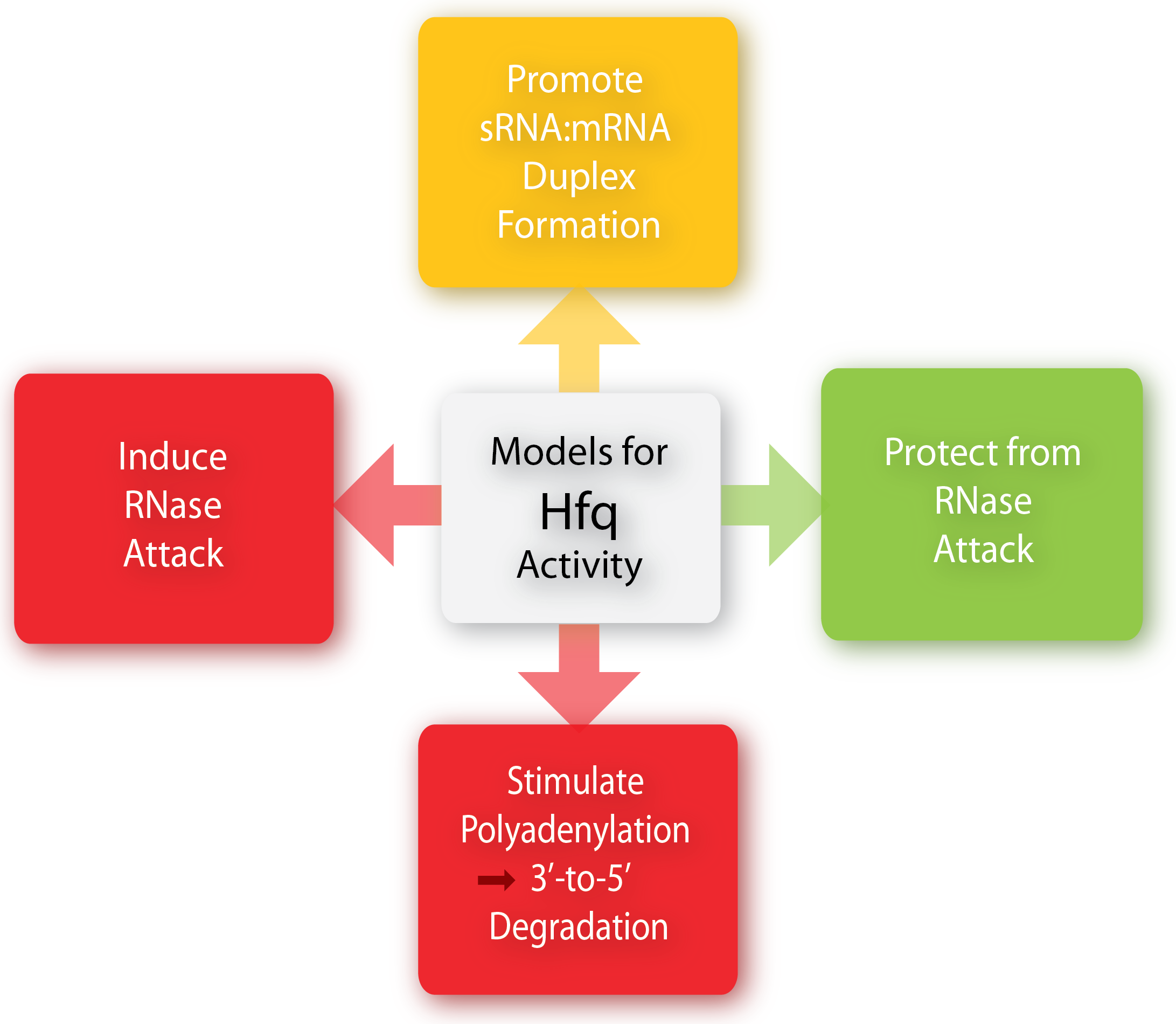
The Hfq chaperone is found to be required for many – but not all – trans-acting antisense sRNAs to form limited base-pairing interactions with their target mRNA[3]. Although Hfq has been the subject of extensive research activity, its specific role, mechanism, and function is still not fully understood and it seems to depend on the specific sRNA-mRNA pair. Several models for the general mechanism of how Hfq mediates regulation and how exactly it facilitates pairing have been proposed and are summarized in a recent paper by Vogel and Luisi, 2011.
Hfq may protect the sRNA from ribonuclease cleavage (often performed by RNase E) in some cases whereas Hfq in other cases induces the cleavage of the sRNA and its target mRNA by RNase E. Furthermore, Hfq may promote the polyadenylation of an mRNA by poly(A) polymerase and hereby triggering the 3’-to-5’ degradation by exoribonucleases[6]. Finally, Hfq can promote duplex formation of the sRNA and mRNA, resulting in either translational repression or initiation, depending on whether the sRNA is an activator or repressor. One way by which Hfq can promote sRNA-mRNA duplex formation is by increasing the on-rate for sRNA annealing to target mRNA by promoting base-pairing, either by inducing changes in the secondary structure of RNA to favor duplex formation or by simultaneously binding both the sRNA and mRNA hereby increasing the local concentration of the two RNAs[6][1].
References
[1] Aiba, Hiroji. “Mechanism of RNA silencing by Hfq-binding small RNAs.” Current Opinion in Microbiology 10, no. 2 (2007): 134-139.
[2] Beisel, Chase L., and Gisela Storz. “Base pairing small RNAs and their roles in global regulatory networks.” FEMS Microbiology Reviews 34, no. 5 (2010): 866-882.
[3] Jousselin, Ambre, Laurent Metzinger, and Brice Felden. “On the facultative requirement of the bacterial RNA chaperone, Hfq.” Trends in Microbiology 17, no. 9 (2009): 399-405.
[4] Levine, Erel, Zhongge Zhang, Thomas Kuhlman, and Terence Hwa. “Quantitative characteristics of gene regulation by small RNA.” PLoS biology. 5, no. 9 (2007).
[5] van Vliet, Arnoud Hm, and Brendan W Wren. “New levels of sophistication in the transcriptional landscape of bacteria.” Genome biology. 10, no. 8 (2009).
[6] Vogel, Jörg, and Ben F Luisi. “Hfq and its constellation of RNA.” Nature reviews. Microbiology 9, no. 8 (2011): 578-589.
[7] Waters, Lauren S., and Gisela Storz. “Regulatory RNAs in Bacteria.” Cell 136, no. 4 (2009): 615-628.
 "
"