Team:EPF-Lausanne/Our Project/T7 promoter variants
From 2011.igem.org
| Line 7: | Line 7: | ||
* [[Team:EPF-Lausanne/Our Project/T7 promoter variants/dnaselect| DNA Selection Experiment]]: Selecting the right DNA | * [[Team:EPF-Lausanne/Our Project/T7 promoter variants/dnaselect| DNA Selection Experiment]]: Selecting the right DNA | ||
* [[Team:EPF-Lausanne/Our Project/T7 promoter variants/t7make| Making T7]]: Making T7 promoter variants for Lysis | * [[Team:EPF-Lausanne/Our Project/T7 promoter variants/t7make| Making T7]]: Making T7 promoter variants for Lysis | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
=== Characterization with RFP === | === Characterization with RFP === | ||
Revision as of 09:06, 21 September 2011
Lysis Selection System
Skip straight to: |
- Platereader Lysis Experiment: Using a platereader
- DNA Recovery Experiment: Recovering DNA after lysis
- DNA Selection Experiment: Selecting the right DNA
- Making T7: Making T7 promoter variants for Lysis
Characterization with RFP
LacI repression
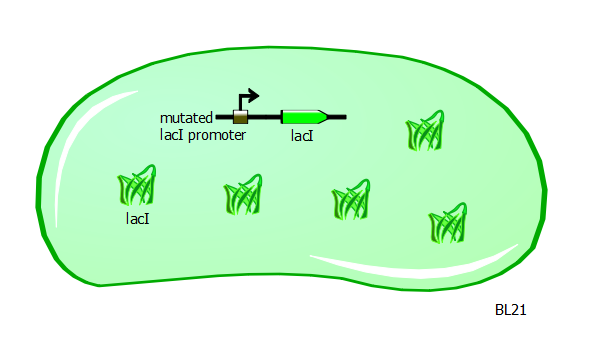
With T7 and T7-lac promoter variants in hand, we want to characterize their relative strengths. To obtain the right set-up, we transform the promoter variant plasmids into a different strain of E. coli called BL21. These cells have a mutation in the promoter for the lacI gene. As a result of this mutation, LacI protein is overproduced and is found abundantly in these cells.
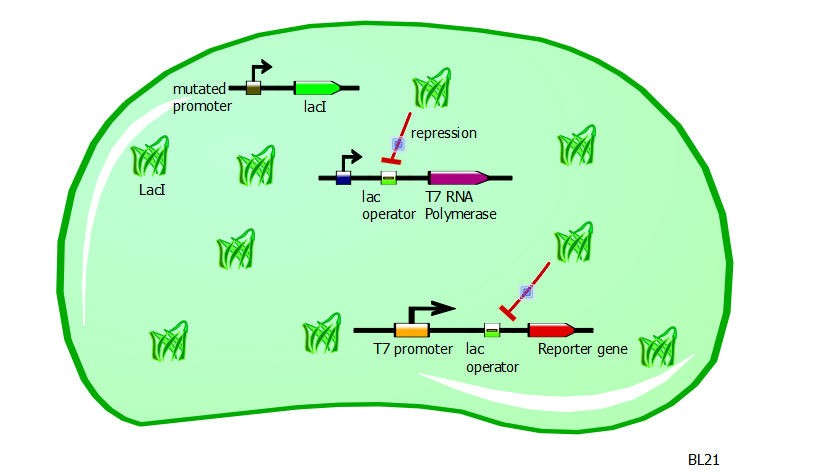
In this same strain, the gene for the T7 RNA polymerase is preceded by a lac operator. Since LacI is a repressor and is strongly present in BL21 cells, the production of T7 RNA polymerases is severely repressed.
With very few T7 RNA polymerases available, there is very little recognition and binding of T7 promoters and consequently very little expression of the gene driven by the T7 promoter. Moreover, the expression of a gene driven by a T7-lac promoter would be even less, since the presence of LacI would block any action of the T7 RNA polymerase.
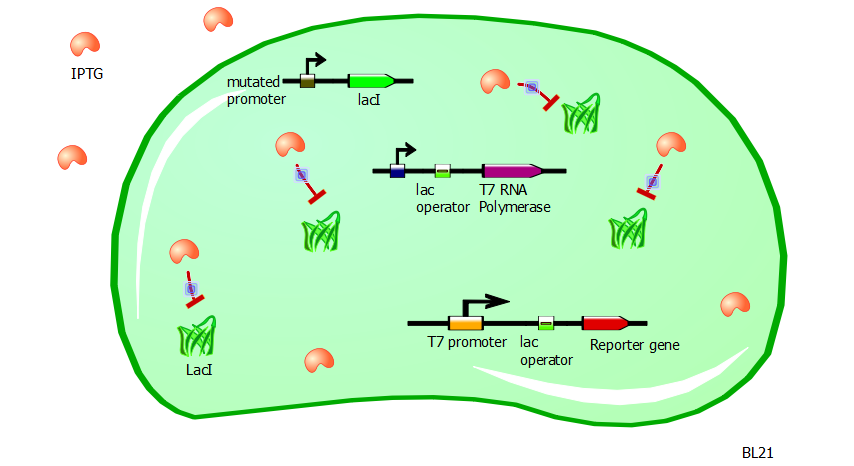
IPTG induction
With such a system in place, the transformation of our T7 and T7-lac promoter variant plasmids into BL21 cells would produce little to no reporter expression. Now to induce promoter activity, we rely on the fact that the chemical IPTG blocks the repressor action of LacI.
Without LacI inhibition, the cell can resume production of T7 RNA polymerases. These in turn can bind to the T7 and T7-lac promoters to express the reporter gene without interruption. So the adding of IPTG to a BL21 cell culture containing these T7 and T7-lac promoter variant plasmids ought to produce high levels of RFP or Lysis expression. Since lysis is a rather binary process (either the cell is lysed or it is not), we use RFP fluorescence as a gauge of promoter strength and efficiency.
DNA Recovery with Lysis
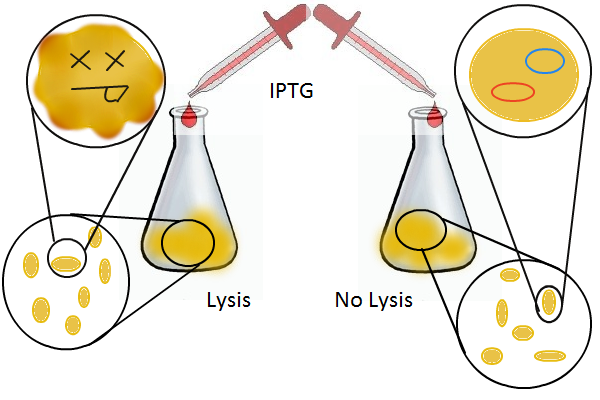
Having a lysis cassette driven by a T7 promoter is an important step towards being able to recover the DNA sequences of transcription factor and promoter mutants that have strong mutual affinities. To verify that basic lysing can be induced, we use the same IPTG experiment as for RFP, using both large samples for visual, qualitative confirmation and small samples in a platereader for quantitative, numerical confirmation.
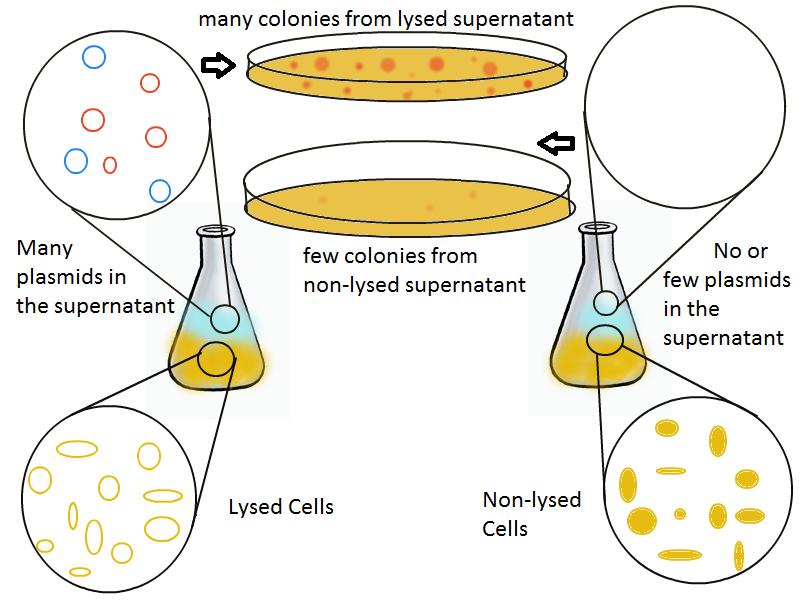
Once the basic mechanism of cell lysing is confirmed, the next step is to show that DNA can be recovered from the supernatant. We grow two large cultures of cells. One contains cells that will lyse and release plasmids into the supernatant while the other has non-lysing, "normal" cells.
Adding IPTG to both flasks induces lysis in one set of cells but not in the others.
Thanks to qPCR, the supernatant harvested from the lysing culture reveals increased numbers of plasmids while the non-lysing culture exhibits significantly lower numbers of plasmids. A similar test involves transforming the supernatant (whose plasmid content is not known a priori) into cells and counting the number of resulting colonies. The plates containing transformations from the lysing supernatant have vastly superior number of colonies compared to the non-lysing supernatant transformations.
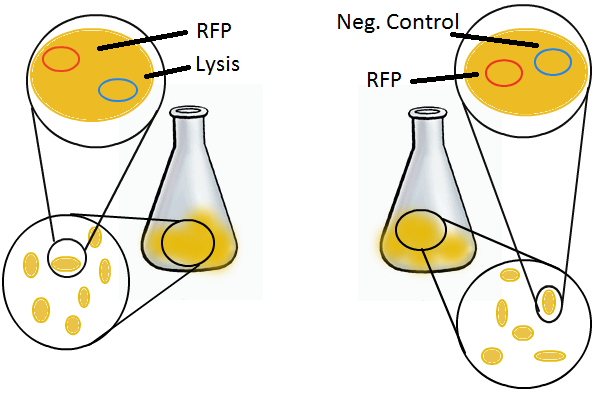
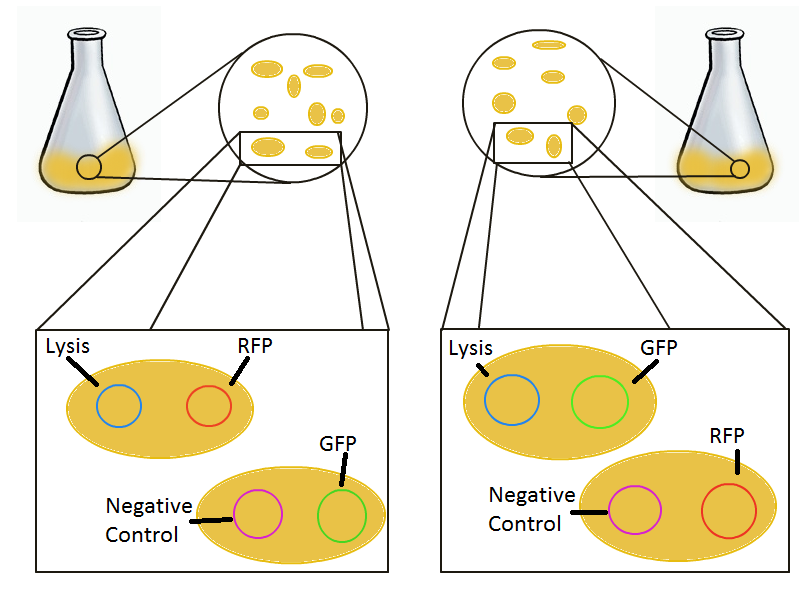
To round out the experiments for DNA recovery, it is essential that we show that the recovered DNA is in fact plasmid DNA from the relevant lysed cells. To that end, we again set up an experiment involving two flasks but this time each flask contains two cultures. For one flask, one culture is a co-transformation of a lysis plasmid and a RFP-containing plasmid and the other is a co-transformation of a negative control plasmid with a GFP-containing plasmid. In the other flask, the reverse is true: lysis is with GFP and negative control is with RFP.
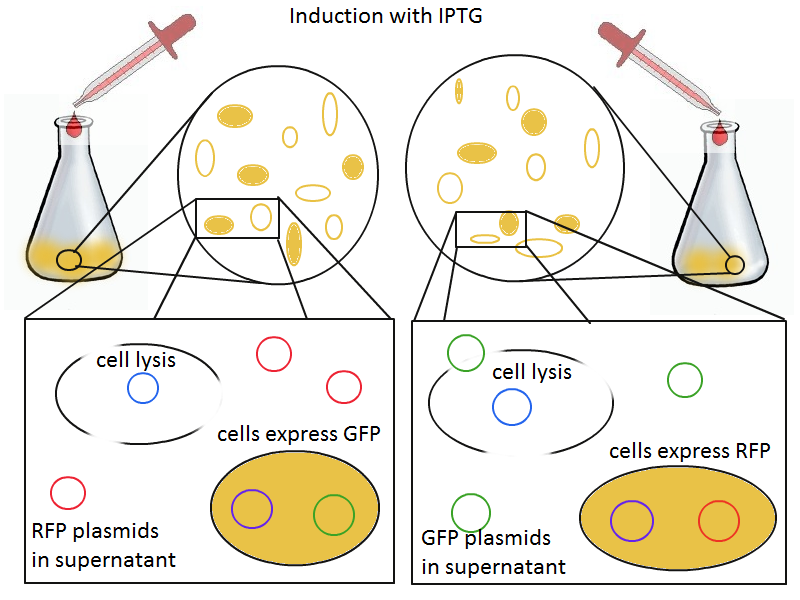
Induction with IPTG lyses the cells with the lysis cassette but leaves the cells with the negative control free to produce the red fluorescence. Collecting the supernatant and sterile filtering it reveals, by qPCR or by transforming it and counting the colonies, that the supernatant contains RFP plasmids in one flask and GFP plasmids in the other. Meanwhile, the large flask from which the supernatant samples are taken should be yellow-green and pink respectively, since the remaining cells will fluoresce according to those colors.
 "
"