Team:NTNU Trondheim/Data
From 2011.igem.org
(Difference between revisions)
(→Stress Sensor) |
|||
| (33 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
= Data = | = Data = | ||
| - | |||
| - | + | ==Our System== | |
| + | [[File:NTNU2011 system.png|center|]] | ||
| - | + | ==Our favorite new parts== | |
| - | + | ||
| - | + | ||
| - | + | #[http://partsregistry.org/Part:BBa_K639003 Parts registry] - '''Stress sensor, BBa_K639003.''' The complete fluorescent stress sensor construct. | |
| - | + | #[http://partsregistry.org/Part:BBa_K639000 Parts registry] - '''Stress sensor precursor, BBa_K639000.''' This brick will yield red colonies as it is. When a promoter is inserted in front of the brick, the red color will disappear or be weaker, due to production of LacI that repress lac-promoter. | |
| - | + | #[http://partsregistry.org/Part:BBa_K639002 Parts registry] - '''rrnB, BBa_K639002.''' rrnB P1 promoter with standard prefix and suffix in BBa format. | |
| - | + | ==Characterization of Pre-existing Parts== | |
| + | #[http://partsregistry.org/Part:BBa_K112118:Experience Parts registry] - '''rrnB P1, BBa_K112118.''' rrnB P1 promoter BioBrick in BBb format made by the Berkley iGEM team in 2008. | ||
| + | #[http://partsregistry.org/Part:BBa_K292006:Experience#User_Reviews Parts registry] - '''LacI repressor, BBa_K292006.''' LacI repressor that SupBiotech-Paris iGEM team made in 2009. | ||
| - | + | ==Other parts== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | #[http://partsregistry.org/Part:BBa_K639001 Parts registry] - '''relA, BBa_K639001.''' relA codes for ppGpp synthase, which has been shown to be useful for regulating the rrnB P1 promoter, mimicking the stringent response when overexpressed. | ||
{{:Team:NTNU_Trondheim/NTNU_footer}} | {{:Team:NTNU_Trondheim/NTNU_footer}} | ||
Latest revision as of 07:37, 21 September 2011

Data
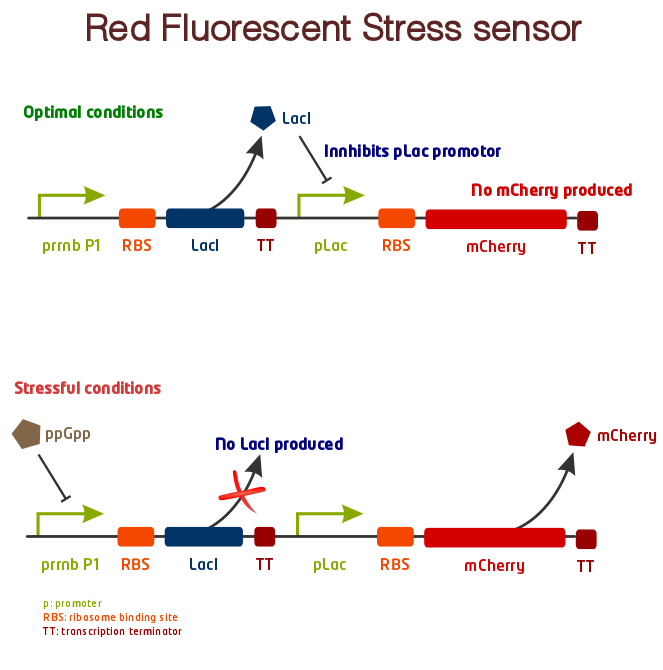
Our System
Our favorite new parts
- [http://partsregistry.org/Part:BBa_K639003 Parts registry] - Stress sensor, BBa_K639003. The complete fluorescent stress sensor construct.
- [http://partsregistry.org/Part:BBa_K639000 Parts registry] - Stress sensor precursor, BBa_K639000. This brick will yield red colonies as it is. When a promoter is inserted in front of the brick, the red color will disappear or be weaker, due to production of LacI that repress lac-promoter.
- [http://partsregistry.org/Part:BBa_K639002 Parts registry] - rrnB, BBa_K639002. rrnB P1 promoter with standard prefix and suffix in BBa format.
Characterization of Pre-existing Parts
- [http://partsregistry.org/Part:BBa_K112118:Experience Parts registry] - rrnB P1, BBa_K112118. rrnB P1 promoter BioBrick in BBb format made by the Berkley iGEM team in 2008.
- [http://partsregistry.org/Part:BBa_K292006:Experience#User_Reviews Parts registry] - LacI repressor, BBa_K292006. LacI repressor that SupBiotech-Paris iGEM team made in 2009.
Other parts
- [http://partsregistry.org/Part:BBa_K639001 Parts registry] - relA, BBa_K639001. relA codes for ppGpp synthase, which has been shown to be useful for regulating the rrnB P1 promoter, mimicking the stringent response when overexpressed.
 "
"