Team:Paris Liliane Bettencourt/Notebook/2011/09/06/
From 2011.igem.org
(→Test of new comptetenc cells) |
|||
| (2 intermediate revisions not shown) | |||
| Line 27: | Line 27: | ||
Positive control with tube one and two are very good, but they are not red!!! How come? | Positive control with tube one and two are very good, but they are not red!!! How come? | ||
| - | ''' | + | ''' To conclude, use the MH1 cells (cap green) for cloning on Cm and Turbo cells to clone on Ampicilyn!!!''' |
| - | To conclude, use the MH1 cells (cap green) for cloning on Cm and Turbo cells to clone on Ampicilyn!!! | + | |
| - | ''' | + | |
=== PCR colony === | === PCR colony === | ||
| Line 36: | Line 34: | ||
An error was made while programming the PCR machine, and the PCR on pHM3 stayed for two hours at 95°C. THe PCR was atempter whatsoever. | An error was made while programming the PCR machine, and the PCR on pHM3 stayed for two hours at 95°C. THe PCR was atempter whatsoever. | ||
| + | |||
| + | == Hovannes-Baptiste == | ||
| + | |||
| + | === Preparation of slides === | ||
| + | |||
| + | Dilution of overnight cultures : YC164 and YC227 (SinOp Design) . <br> | ||
| + | |||
| + | |||
| + | YC227 has a hyperspank promoter (inducible by IPTG) which regulates a GFP gene. It also produces phosphorilated KinA.<br> | ||
| + | YC164 Contains SpoA which can be indirectly phosphorilated by KinA. Phosphorilated SpoA indirectly activates a gene which contains GPF and SinI. SinI is a signal involved in biofilm shaped growth.<br> | ||
| + | |||
| + | This time, we waited to an OD of 0.6 (600 nm) rather than 0.4 to see if it was more suited to our needs. | ||
| + | |||
| + | Two well slides : 1-control (YC164 with IPTG) 2-Mix (both strains with IPTG) | ||
| + | <br> | ||
| + | |||
| + | === Observation === | ||
| + | -37°C Microscopy-<br> | ||
| + | |||
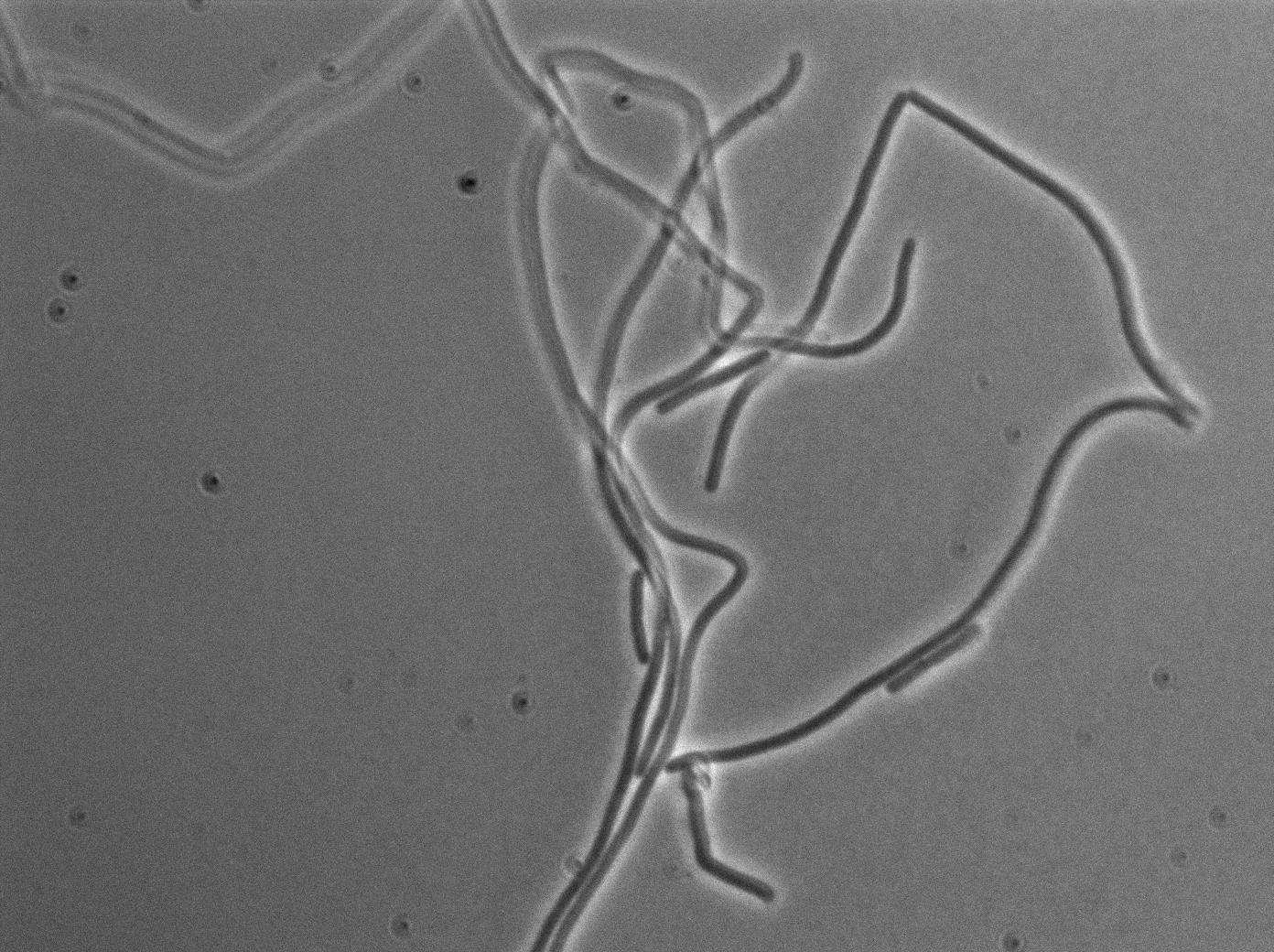
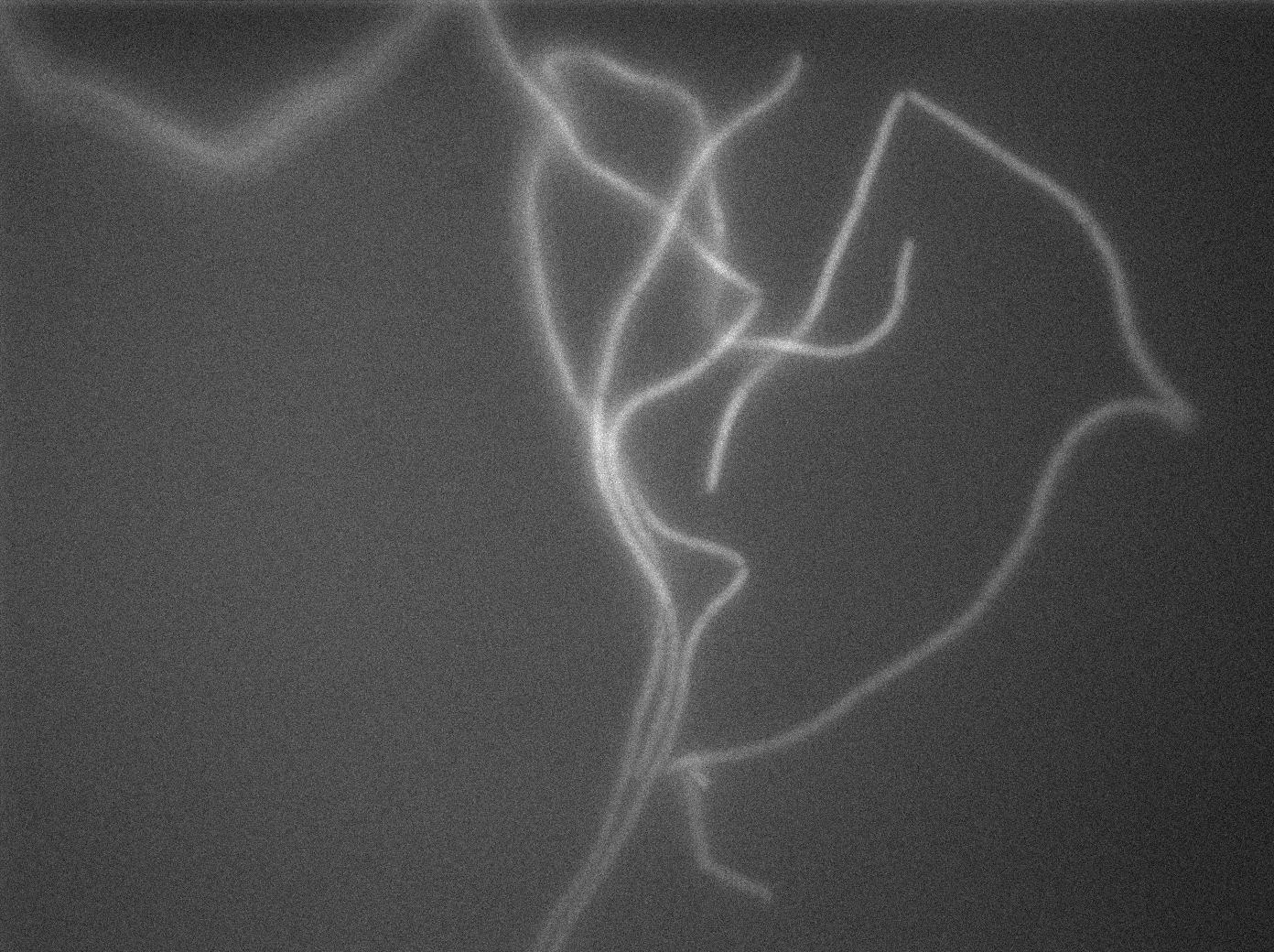
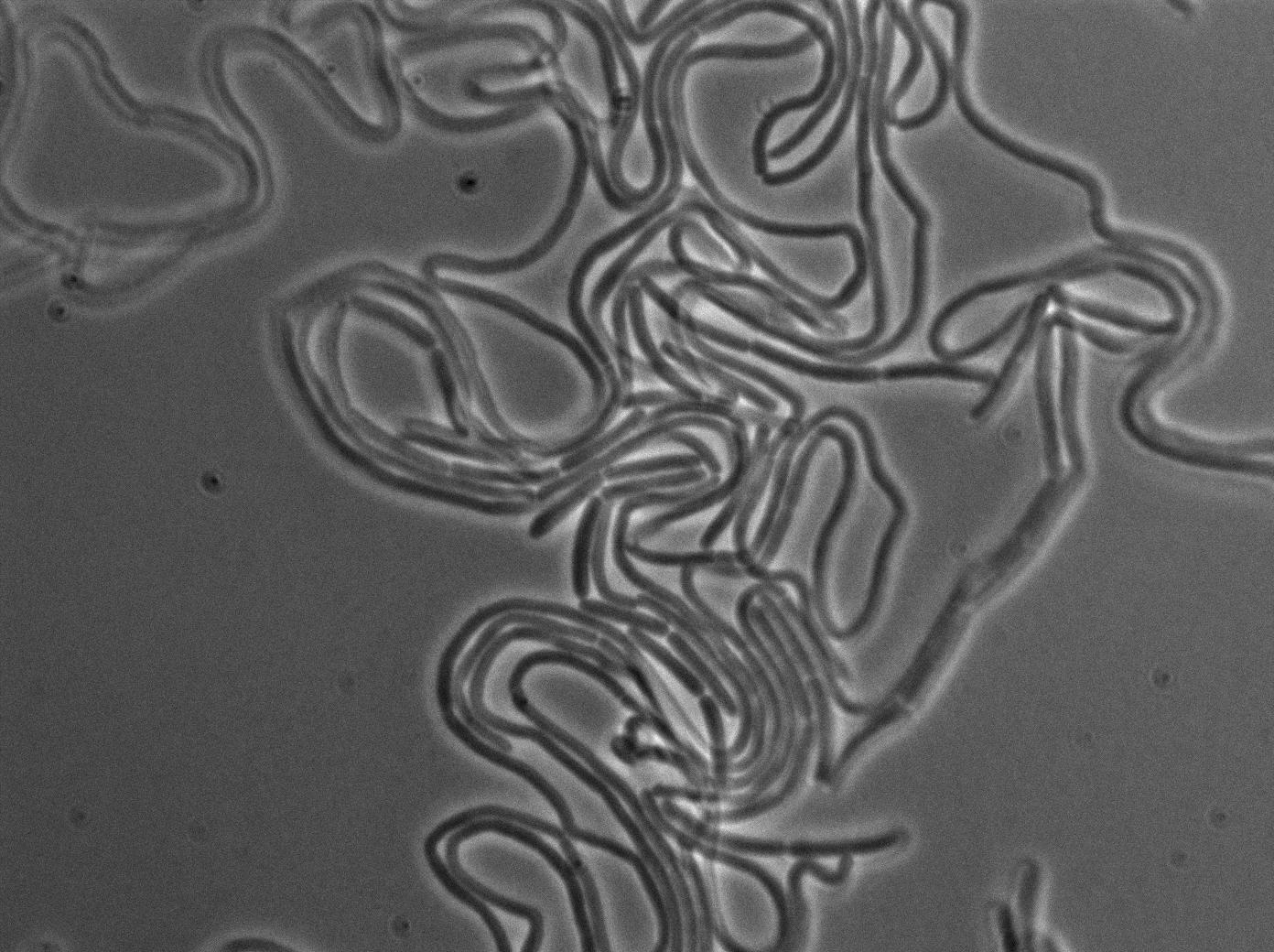
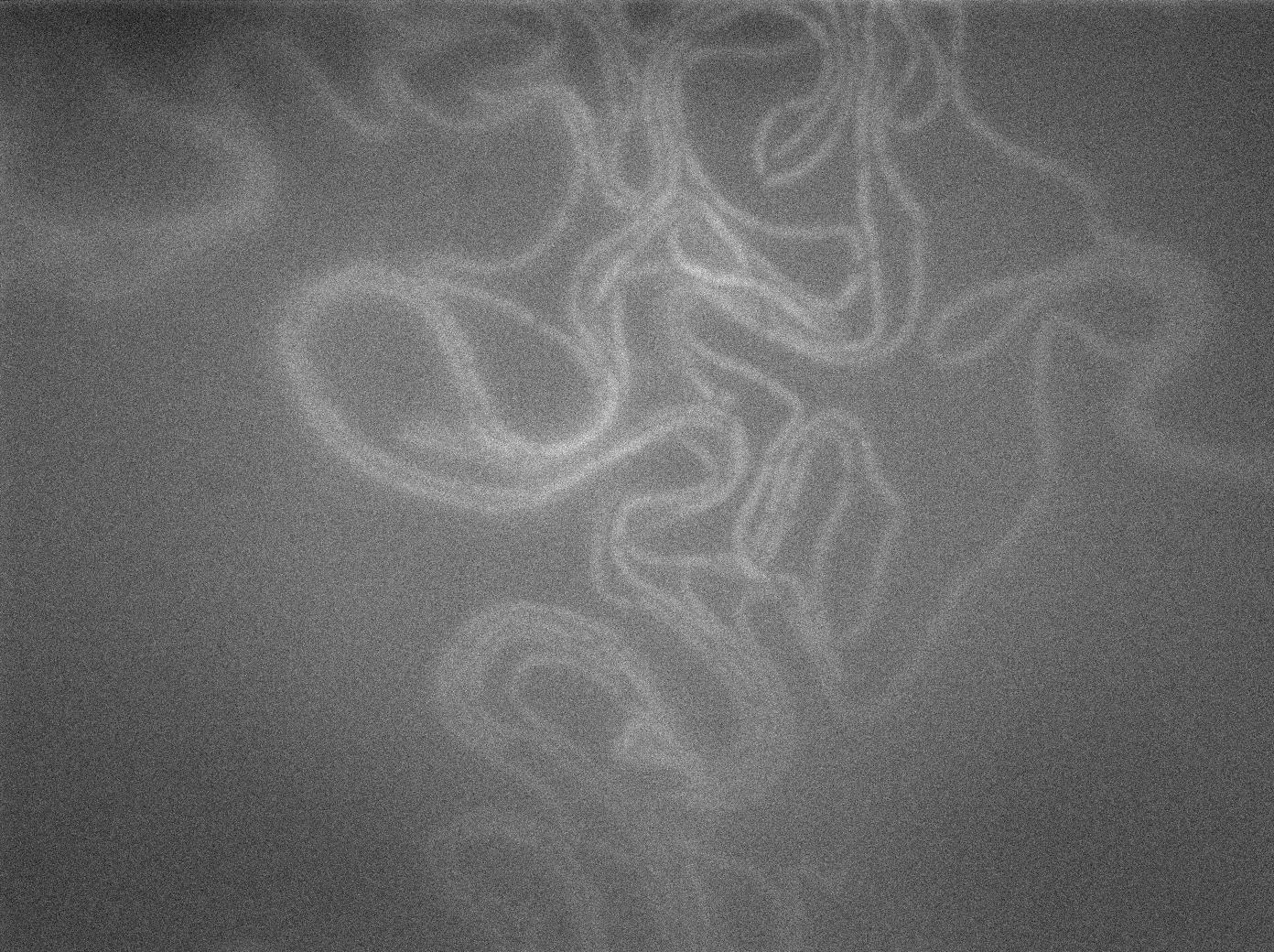
| + | We observed the plate with TRANS and YFP-filter settings on the Old Zeiss. Unfortunately, this microscope gets quickly out of focus so we need to take each picture manually. As expected, YC227 was producing GFP at the begining of the experiment, while YC164 was not. Our observations over 4 hours are the following: | ||
| + | *Both strains seem to grow at approximately the same rate (which means more GFP dilution and less clear GFP images) | ||
| + | *We do not see any evidence of nanotube yet. Some image enhancing might be required in the next days on our results. | ||
| + | |||
| + | [[File:0906_SinOp_trans1.jpg|500px|thumb|center|TRANS at t=0min]] | ||
| + | [[File:0906_SinOp_fluo1.jpg|500px|thumb|center|GFP at t=0min]] | ||
| + | [[File:0906_SinOp_trans24.jpg|500px|thumb|center|TRANS at t=180min]] | ||
| + | [[File:0906_SinOp_fluo24.jpg|500px|thumb|center|GFP at t=180min]] | ||
| + | |||
| + | <html> | ||
| + | <!-- PAGE FOOTER -- ITEMS FROM COLUMN ! HAVE BEEN MOVED HERE -- RDR --> | ||
| + | |||
| + | <div id="footer-wrapper"> | ||
| + | <div id="footer"> | ||
| + | <div id="f-poweredbyico"><a href="http://www.mediawiki.org/"><img src="/wiki/skins/common/images/poweredby_mediawiki_88x31.png" height="31" width="88" alt="Powered by MediaWiki" /></a></div> <div id="f-copyrightico"><a href="http://creativecommons.org/licenses/by/3.0/"><img src="http://i.creativecommons.org/l/by/3.0/88x31.png" alt="Attribution 3.0 Unported" width="88" height="31" /></a></div> <ul id="f-list"> | ||
| + | |||
| + | |||
| + | <!-- Recentchanges is not handles well DEBUG --> | ||
| + | <li id="t-recentchanges"><a href="/Special:RecentChanges" | ||
| + | title='Recent changes'>Recent changes</a></li> | ||
| + | |||
| + | <li id="t-whatlinkshere"><a href="/Special:WhatLinksHere/Team:Paris_Bettencourt/Modeling" | ||
| + | title="List of all wiki pages that link here [j]" accesskey="j">What links here</a></li> | ||
| + | |||
| + | <li id="t-recentchangeslinked"><a href="/Special:RecentChangesLinked/Team:Paris_Bettencourt/Modeling" | ||
| + | title="Recent changes in pages linked from this page [k]" accesskey="k">Related changes</a></li> | ||
| + | |||
| + | |||
| + | |||
| + | <li id="t-upload"><a href="/Special:Upload" | ||
| + | title="Upload files [u]" accesskey="u">Upload file</a> | ||
| + | </li> | ||
| + | <li id="t-specialpages"><a href="/Special:SpecialPages" | ||
| + | title="List of all special pages [q]" accesskey="q">Special pages</a> | ||
| + | |||
| + | </li> | ||
| + | <li><a href='/Special:Preferences'>My preferences</a></li> | ||
| + | </ul> | ||
| + | </div> <!-- close footer --> | ||
| + | <div id='footer'> | ||
| + | <ul id="f-list"> | ||
| + | |||
| + | <li id="t-print"><a href="/wiki/index.php?title=Team:Paris_Bettencourt/Modeling&printable=yes" | ||
| + | title="Printable version of this page [p]" accesskey="p">Printable version</a> | ||
| + | |||
| + | </li> | ||
| + | |||
| + | <li id="t-permalink"><a href="/wiki/index.php?title=Team:Paris_Bettencourt/Modeling&oldid=86565" | ||
| + | title="Permanent link to this revision of the page">Permanent link</a> | ||
| + | </li> | ||
| + | |||
| + | |||
| + | <li id="privacy"><a href="/2011.igem.org:Privacy_policy" title="2011.igem.org:Privacy policy">Privacy policy</a></li> | ||
| + | <li id="disclaimer"><a href="/2011.igem.org:General_disclaimer" title="2011.igem.org:General disclaimer">Disclaimers</a></li> | ||
| + | </ul> | ||
| + | |||
| + | </div> <!-- close footer --> | ||
| + | </div> <!-- close footer-wrapper --> | ||
| + | </div> | ||
| + | </html> | ||
Latest revision as of 13:53, 9 September 2011

Contents |
Cyrille
Transformations check
pVeg-YFP in pHM3: Transformation works. About the same ratio in 1:1 and 1:10. A bit less in 1:100.
-> Transformation seems clean, control weak but correct
-> 10 colonies insulated
TetO-pVeg-YFP in pHM3: Transformation works, a bit less eficiency. It seems clean. (no negative control)
-> A bit weaker, control correct
-> 10 colonies insulated
YFP TetR BB in pHM3: Negative control very polluted. About 1.3 times the number of colonies in the T1 and very few in the T2. Positive control correct.
-> 10 colonies insulated from plate T1
ComS in pSB1C3: Negative control very clean (~20 colonies in the 200µL plate) Very few colonies on the other plates. Seems to have worked best with ratio 1:10-5. Very fw colonies on the other plates
-> 10 colonies insulated
Test of new comptetent cells
Negative control on Ampicylin are clean Negative control (400µL out of 1ml plated)on Cm are a bit polluted. About 30 clones.
Positive control with tube one and two are very good, but they are not red!!! How come?
To conclude, use the MH1 cells (cap green) for cloning on Cm and Turbo cells to clone on Ampicilyn!!!
PCR colony
The clones insulated was also PCR colonysed. We used a Taq MasterMix.
An error was made while programming the PCR machine, and the PCR on pHM3 stayed for two hours at 95°C. THe PCR was atempter whatsoever.
Hovannes-Baptiste
Preparation of slides
Dilution of overnight cultures : YC164 and YC227 (SinOp Design) .
YC227 has a hyperspank promoter (inducible by IPTG) which regulates a GFP gene. It also produces phosphorilated KinA.
YC164 Contains SpoA which can be indirectly phosphorilated by KinA. Phosphorilated SpoA indirectly activates a gene which contains GPF and SinI. SinI is a signal involved in biofilm shaped growth.
This time, we waited to an OD of 0.6 (600 nm) rather than 0.4 to see if it was more suited to our needs.
Two well slides : 1-control (YC164 with IPTG) 2-Mix (both strains with IPTG)
Observation
-37°C Microscopy-
We observed the plate with TRANS and YFP-filter settings on the Old Zeiss. Unfortunately, this microscope gets quickly out of focus so we need to take each picture manually. As expected, YC227 was producing GFP at the begining of the experiment, while YC164 was not. Our observations over 4 hours are the following:
- Both strains seem to grow at approximately the same rate (which means more GFP dilution and less clear GFP images)
- We do not see any evidence of nanotube yet. Some image enhancing might be required in the next days on our results.
 "
"