Team:EPF-Lausanne/Notebook/Summary
From 2011.igem.org
(Project overview) |
|||
| (5 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | = | + | {{:Team:EPF-Lausanne/Templates/Header|title=Our Project}} |
For this year’s iGEM competition our team has set out to find a method for producing new regulatory parts used in the construction of complex biological devices. | For this year’s iGEM competition our team has set out to find a method for producing new regulatory parts used in the construction of complex biological devices. | ||
| Line 8: | Line 8: | ||
Engineering new transcription factors will not only give us a better idea of how they work but also extend the tool kit of the synthetic biologist. Within synthetic biology, TFs are used to design biological circuits where they act as building blocks for reporters, inverters, logic functions and switches . Extending the tool kit with new TFs will allow combinations of the devices that today share the same regulatory elements and thus cannot be used in the same system. | Engineering new transcription factors will not only give us a better idea of how they work but also extend the tool kit of the synthetic biologist. Within synthetic biology, TFs are used to design biological circuits where they act as building blocks for reporters, inverters, logic functions and switches . Extending the tool kit with new TFs will allow combinations of the devices that today share the same regulatory elements and thus cannot be used in the same system. | ||
| + | |||
| + | [[File:EPFL-WhiteboardPlasmids.JPG|650px]] | ||
| + | |||
Our project’s outcome will be a method to derive pairs of TFs and DNA binding sites meeting selection criteria for orthogonality, affinity and specificity. It will include setting a selection scheme that allows for high throughput testing identification of functional TF-consensus sequence pairs in vitro and in vivo. We plan to use bacterial culture in parallel chemostat platforms and MITOMI to screen a library of TFs against a set of consensus sequences. | Our project’s outcome will be a method to derive pairs of TFs and DNA binding sites meeting selection criteria for orthogonality, affinity and specificity. It will include setting a selection scheme that allows for high throughput testing identification of functional TF-consensus sequence pairs in vitro and in vivo. We plan to use bacterial culture in parallel chemostat platforms and MITOMI to screen a library of TFs against a set of consensus sequences. | ||
| Line 14: | Line 17: | ||
We want to enhance the diversity of the repressor-TF class by engineering new transcription factors that can bind unique promoter sequences different from the wild-type DNA binding site normally associated with TetR. Our proposed research plan for this summer is as follows: | We want to enhance the diversity of the repressor-TF class by engineering new transcription factors that can bind unique promoter sequences different from the wild-type DNA binding site normally associated with TetR. Our proposed research plan for this summer is as follows: | ||
| + | |||
1) introduce mutations in the TetR sequence and its binding site | 1) introduce mutations in the TetR sequence and its binding site | ||
| + | |||
2) identify promising TF-binding site pairs (using in vivo and in vitro techniques) | 2) identify promising TF-binding site pairs (using in vivo and in vitro techniques) | ||
| + | |||
3) conduct multiple rounds of selection to improve affinity and specificity to optimize new pairs of repressor-TFs. | 3) conduct multiple rounds of selection to improve affinity and specificity to optimize new pairs of repressor-TFs. | ||
| + | |||
| + | {{:Team:EPF-Lausanne/Templates/Footer}} | ||
Latest revision as of 10:00, 7 June 2011
Our Project
For this year’s iGEM competition our team has set out to find a method for producing new regulatory parts used in the construction of complex biological devices.
Transcription is the first step of gene expression that can be regulated. Key elements of this regulation are proteins recognising and binding specific DNA sequences that either recruit the full transcription machinery or prevent it from assembling on the DNA. It is this class of proteins, called transcription factors (TFs), that we intend to engineer.
Existing DNA binding proteins can be a starting point for developing these new TFs. By changing its residues it is possible to modify the affinity of these proteins to a specific binding site and interact with the activation or repression mechanisms of the downstream gene transcription. Investigating these effects can improve our understanding of how the residue composition affects the TF's ability to recognise a given DNA consensus sequence.
Engineering new transcription factors will not only give us a better idea of how they work but also extend the tool kit of the synthetic biologist. Within synthetic biology, TFs are used to design biological circuits where they act as building blocks for reporters, inverters, logic functions and switches . Extending the tool kit with new TFs will allow combinations of the devices that today share the same regulatory elements and thus cannot be used in the same system.
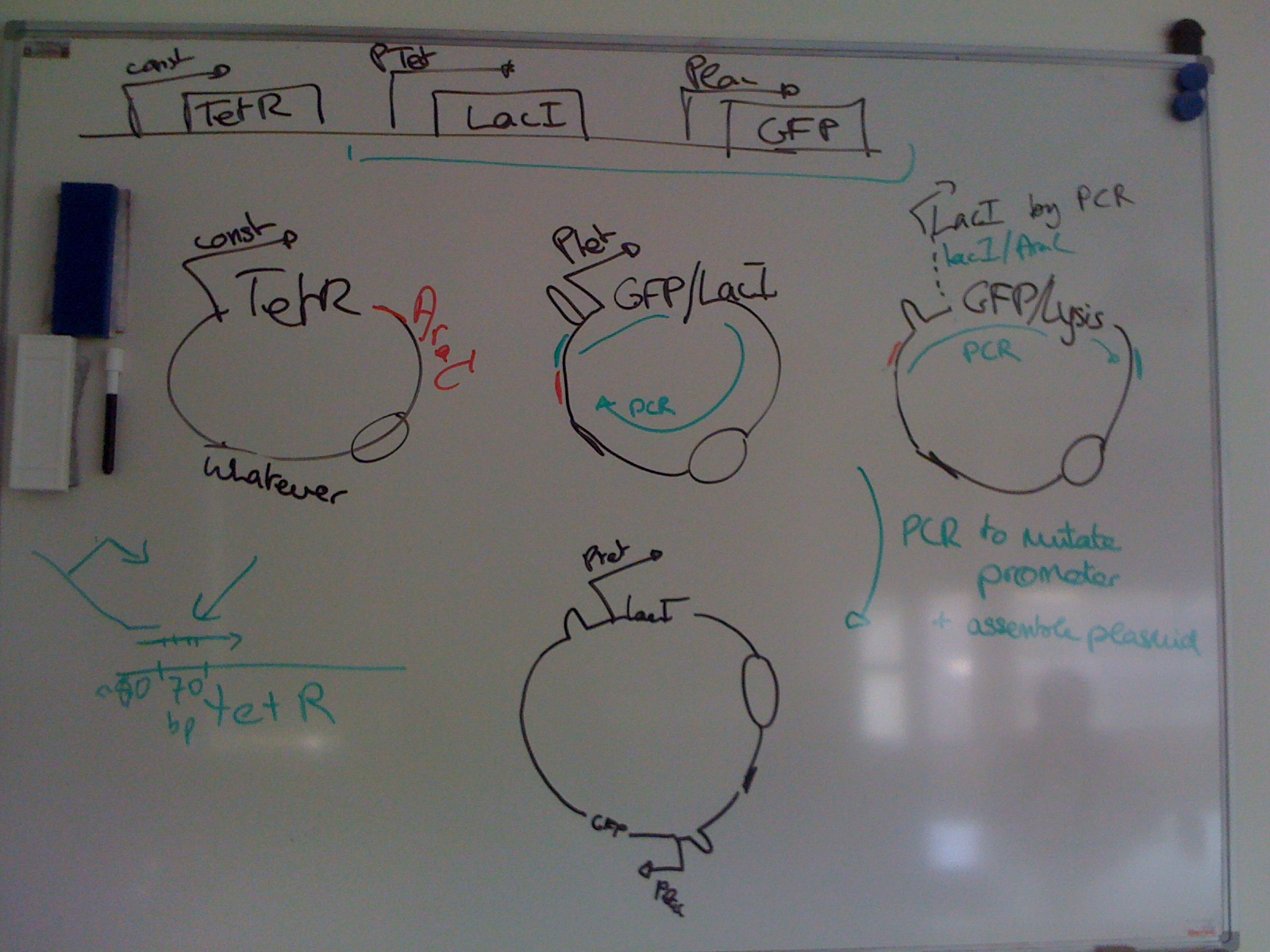
Our project’s outcome will be a method to derive pairs of TFs and DNA binding sites meeting selection criteria for orthogonality, affinity and specificity. It will include setting a selection scheme that allows for high throughput testing identification of functional TF-consensus sequence pairs in vitro and in vivo. We plan to use bacterial culture in parallel chemostat platforms and MITOMI to screen a library of TFs against a set of consensus sequences.
Our research will begin with the tetracycline repressor (TetR), a transcription factor which down-regulates (represses) the transcription of the adjacent gene. This protein and its mutants are often used by synthetic biologists “as control elements to regulate gene expression” due to their detailed characterisation, but also because of the limited number of proteins available for use in the repressor-TF class.
We want to enhance the diversity of the repressor-TF class by engineering new transcription factors that can bind unique promoter sequences different from the wild-type DNA binding site normally associated with TetR. Our proposed research plan for this summer is as follows:
1) introduce mutations in the TetR sequence and its binding site
2) identify promising TF-binding site pairs (using in vivo and in vitro techniques)
3) conduct multiple rounds of selection to improve affinity and specificity to optimize new pairs of repressor-TFs.
 "
"