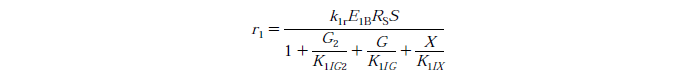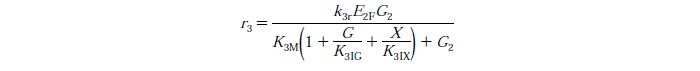Team:Edinburgh/Cellulases (MATLAB model)
From 2011.igem.org
| Line 25: | Line 25: | ||
===Rate Equations=== | ===Rate Equations=== | ||
| - | [[File:Edinburgh-Rate-1.png|thumb|left|700px|Cellulose to Cellobiose Reaction with Competitive Glucose, Cellobiose and Xylose Inhibition. | + | [[File:Edinburgh-Rate-1.png|thumb|left|700px|Cellulose to Cellobiose Reaction with Competitive Glucose, Cellobiose and Xylose Inhibition.]] |
| + | |||
| + | [[File:Edinburgh-Rate-2.png|thumb|left|700px|Cellulose to Glucose Reaction with Competitive Glucose, Cellobiose and Xylose Inhibition.]] | ||
| + | |||
| + | [[File:Edinburgh-Rate-3.png|thumb|left|700px|Cellobiose to Glucose Reaction with competitive Glucose, Cellibiose and Xylose Inhibition.]] | ||
| + | |||
| + | |||
| + | '''Constants''' | ||
k1r is the reaction rate constant for reaction 1 | k1r is the reaction rate constant for reaction 1 | ||
E1B is the bound concentration for exo and endo-beta-1,4-glucanase<br/> | E1B is the bound concentration for exo and endo-beta-1,4-glucanase<br/> | ||
| Line 35: | Line 42: | ||
K1IG is the inhibition constant for Glucose at reaction 1<br/> | K1IG is the inhibition constant for Glucose at reaction 1<br/> | ||
K1IX is the xylose inhibition constant for reaction 1.<br/> | K1IX is the xylose inhibition constant for reaction 1.<br/> | ||
| - | Note: Assuming no xylose inhibition therefore X=0 | + | Note: Assuming no xylose inhibition therefore X=0 |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Revision as of 13:37, 6 September 2011
Cellulases (MATLAB model)
The most important part of the biorefinery is the reactor where cellulose is converted to glucose. But accurately predicting how much is converted, using synergy between enzymes is difficult without literature to provide the ordinary differential equations (ODE's) and the kinetic parameters. Therefore this model only looks at the free floating enzyme approach (non-synergy). It is deterministic i.e non random and is set by a series of initial conditions.
Contents |
Assumptions
The mathematical model is based on the ODE's and kinetic parameters outlined in [http://onlinelibrary.wiley.com/doi/10.1021/bp034316x/full Kadam et al, 2004]. The following are its assumptions and basis:
- Rate equations assume enzyme adsorption follows the Langmuir isotherm model
- Glucose and cellobiose which are the products of cellulose hydrolysis, were assumed to, 'competitively inhibit enzyme hyrolysis' [http://onlinelibrary.wiley.com/doi/10.1021/bp034316x/full Kadam et al, 2004]
- Assume all reactions follow the same temperature dependency Arrhenius relationship. However it should be different for every enzyme component, 'because of their varying degrees of thermostability, with beta-glucocidase being the most thermostable. Hence the assumption is a simplification of reality' [http://onlinelibrary.wiley.com/doi/10.1021/bp034316x/full Kadam et al, 2004]
- Conversion of cellobiose to glucose follows the Michaelis-Menten enzyme kinetic model
Equations
Rate Equations
Constants
k1r is the reaction rate constant for reaction 1
E1B is the bound concentration for exo and endo-beta-1,4-glucanase
Rs is a substrate reactivity parameter, S is the substrate reactivity at a given time
G2 is te concentration of cellobiose
G is the concentration of Glucose
X is the xylose concentration
K1IG2 is the inhibition constant for cellobiose at reaction 1
K1IG is the inhibition constant for Glucose at reaction 1
K1IX is the xylose inhibition constant for reaction 1.
Note: Assuming no xylose inhibition therefore X=0
References
- Kadam KL, Rydholm EC, McMillan JD (2004) [http://onlinelibrary.wiley.com/doi/10.1021/bp034316x/full Development and Validation of a Kinetic Model for Enzymatic Saccharification of Lignocellulosic Biomass]. Biotechnology Progress 20(3): 698–705 (doi: 10.1021/bp034316x).
 "
"