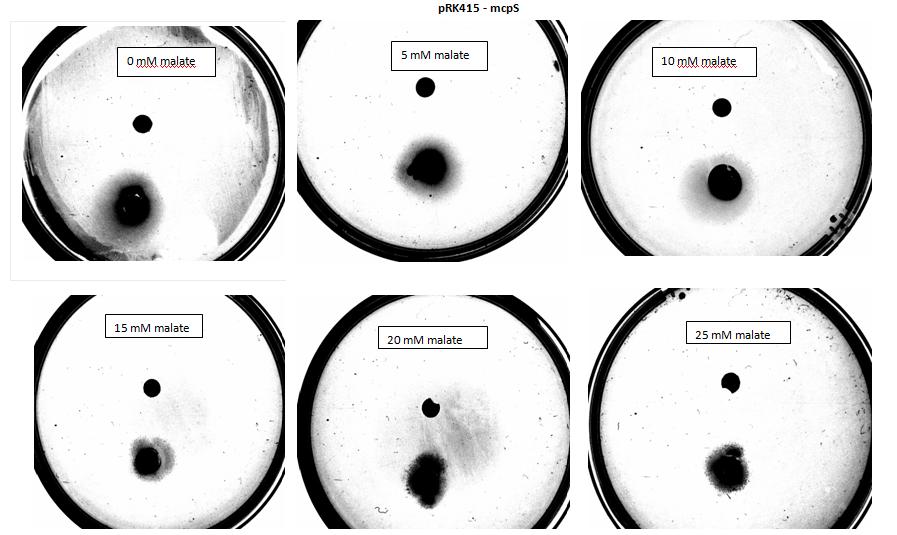
Team:Imperial College London/Project/Chemotaxis/Results
From 2011.igem.org
| Line 22: | Line 22: | ||
<p>The chemotaxis pathway in E.Coli is demonstrated in Figure 1. MCPs form stable ternary complexes with the CheA and CheW proteins to generate signals that control the direction of rotation of the flagellar motors [5]. The signaling currency is in the form of phosphoryl groups (p), made available to the CheY and CheB effector proteins through autophosphorylation of CheA[1]. CheYp initiates flagellar responses by interacting with the motor to enance the probability of ‘run’[1]. CheBp is part of a sensory adaptation circuit that terminates motor responses[1]. MCP complexes have two alternative CheA autokinase activity; When the receptor is not occupied by chemoattractant, the receptor stimulates CheA activity[1]. The overall flux of phosphoryl groups to inhibited and stimulated states. Changes in attractant concentration shift this distribution, triggering a flagellar response [1]. The ensuing changes in CheB phosphorylation state alter its methylesterase activity, producing a net change in MCP methylation state that cancels the stimulus signal [1]. </P> | <p>The chemotaxis pathway in E.Coli is demonstrated in Figure 1. MCPs form stable ternary complexes with the CheA and CheW proteins to generate signals that control the direction of rotation of the flagellar motors [5]. The signaling currency is in the form of phosphoryl groups (p), made available to the CheY and CheB effector proteins through autophosphorylation of CheA[1]. CheYp initiates flagellar responses by interacting with the motor to enance the probability of ‘run’[1]. CheBp is part of a sensory adaptation circuit that terminates motor responses[1]. MCP complexes have two alternative CheA autokinase activity; When the receptor is not occupied by chemoattractant, the receptor stimulates CheA activity[1]. The overall flux of phosphoryl groups to inhibited and stimulated states. Changes in attractant concentration shift this distribution, triggering a flagellar response [1]. The ensuing changes in CheB phosphorylation state alter its methylesterase activity, producing a net change in MCP methylation state that cancels the stimulus signal [1]. </P> | ||
| - | <p><img src="https://static.igem.org/mediawiki/2011/ | + | <p><img src="https://static.igem.org/mediawiki/2011/d/d7/chemotaxis pathway.png" alt="" width="131" height="29" /> </p> |
| + | <p>'''Figure 1[1]: Chemotaxis signaling components and pathways for E.Coli.'''</p> | ||
| + | |||
Revision as of 15:03, 4 September 2011
Chemotaxis Results
Modelling
Introduction
E.coli is a motile strain of bacteria, which is to say it can swim. It is able to do so by rotating its flagellum, which is a rotating tentacle like structure on the outside of cell. Chemotaxis is the movement up concentration gradient of chemoattractants (i.e. malate in our project) and away from poisons. E.coli is too small to detect any concentration gradient between the two ends of itself, and so they must randomly head in any direction and then compare the new chemoattractant concentration at new point to the previous 3-4s point. Its motion is described by ‘runs’ and ‘tumbles’, runs refer to a smooth, straight line movement for a number of seconds, while tumble referring to reorientation of bacteria [1]. Chemoattractant increases transiently raise the probability of ‘tumble’ (or bias), and then a sensory adaptation process returns the bias to baseline, enabling the cell to detect and respond to further concentration changes. The response to a small step change in chemoattractant concentration in a spatially uniform environment increase the response time occurs over a 2- to 4- s time span [2]. Saturating changes in chemoattractant can increase the response time to several minutes.
Many bacterial chemoreceptors belong to a family of transmemberane methyl-accepting chemotaxis proteins (MCPs) [3]. Each chemoreceptors on the bacterium has a periplasmic binding domain and a cytoplasmic signaling domain that communicates with the flagellar motors via a phosphorelay sequence involving the CheA, CheY, and CheZ proteins. Modeling of this chemotaxis pathway in single cell is very important, as it will help us to determine, with certain numbers of chemoreceptors, the threshold of chemoattractant concentration where the bacterium is able to detect and the saturation level of chemoattractant where the all the receptors on the bacterium are occupies. A it is believed that the auxin should be kept at a very close region around the root (0.25 cm [4]), therefore it is very important to obtain the number of chemoreceptors needed on individual bacterium that enables the it to stay close to the plant root/seed from modeling.
In addition, modeling of chemotaxis of bacteria population is also valuable for us to capture the overview of movement of bacteria around the plant root; therefore it can potentially inform our project about how and where we can place our bacteria. Under experiment condition, the chemoattractant diffuses all the time from the source. However, in real soil, the root produces malate all the time, therefore we assume that the distribution of chemoattractant outside the root is steady and time-independent. Hence, the modeling of bacteria population chemotaxis will be built with different patterns of chemoattractant distributions.
Chemotaxis Pathway
The chemotaxis pathway in E.Coli is demonstrated in Figure 1. MCPs form stable ternary complexes with the CheA and CheW proteins to generate signals that control the direction of rotation of the flagellar motors [5]. The signaling currency is in the form of phosphoryl groups (p), made available to the CheY and CheB effector proteins through autophosphorylation of CheA[1]. CheYp initiates flagellar responses by interacting with the motor to enance the probability of ‘run’[1]. CheBp is part of a sensory adaptation circuit that terminates motor responses[1]. MCP complexes have two alternative CheA autokinase activity; When the receptor is not occupied by chemoattractant, the receptor stimulates CheA activity[1]. The overall flux of phosphoryl groups to inhibited and stimulated states. Changes in attractant concentration shift this distribution, triggering a flagellar response [1]. The ensuing changes in CheB phosphorylation state alter its methylesterase activity, producing a net change in MCP methylation state that cancels the stimulus signal [1].
![]()
'''Figure 1[1]: Chemotaxis signaling components and pathways for E.Coli.'''
Malate concentration distribution
For our project, malate is the chemoattractant that results in the movement of E.coli. In this section, we will first model the concentration distribution of the chemoattractant, malate in the soil. Then, we will model the bacteria concentration pattern as a result of this distribution of malate. Finally, we will infer some useful information by analysing the results of the modelling.
We will model the concentration distributions of malate and bacteria using the Keller-Segel model which is governed by the two equations shown below. Solving the equations will give the concentration distributions of the malate and the bacteria respectively
 ---------------------------------------------------------------------------------------------(1)
---------------------------------------------------------------------------------------------(1)
 -------------------------------------------------------(2)
-------------------------------------------------------(2)
s = concentration of chemoattractant
D = diffusion coefficient of chemoattractant
f = degradation of chemoattractant
b = number concentration of bacteria
µ = bacterial diffusion coefficient (how fast bacteria spread)
χ = chemotactic coefficient (how sensitive bacteria are)
g = bacterial cell growth
h = bacterial cell death
The values of the above parameters for E. coli are shown in the following table. These values will be used for the modelling.
Parameter description |
Notation |
Value |
Initial bacterial concentration |
b0 |
108 cells/ml |
Initial attractant concentration |
s0 |
0.1 mM or 0.1 mol/m3 |
Bacterial diffusion coefficient |
µ |
1.5*10-5 cm2/s |
Bacterial chemotactic coefficient |
χ |
1.5-75*10-5 cm2/s |
Attractant diffusion coefficient |
D |
10-5 cm2/s |
Reference: Overview of Mathematical Approaches Used to Model Bacterial Chemotaxis II: Bacterial Populations
The assumptions that we have made are as follow:
- The entire root system is assumed to take the shape of a long cylinder. Hence, a cylindrical coordinate system will be used.
- The system is axisymmetric and there is no variation along the vertical length of the root. Hence,

- The system has reached steady state and is time-independent. Hence,

- Degradation of chemoattractant is first order and is described by f = ks where k is the degradation rate of the chemoattractant.
- Bacterial cell growth and death are neglected. Hence, g(b,s) = h(b,s) = 0
Applying the assumptions above, equation (1) becomes
![]()
Rearranging,
![]() -------------------------------------------------(3)
-------------------------------------------------(3)
Equation (3) is in the form of the modified Bessel equation. Hence, the solution of equation (3) is given by,
![]()
Where K0 is the modified Bessel function.
Since it is unrealistic for the concentration to increase to infinity, A=0. And applying the boundary condition, the solution becomes,
![]() -----------------------------------------------------(4)
-----------------------------------------------------(4)
The modeling result show that the steady-state pattern of malate distribution. The concentration of malate is a variable against the distance


Chemotaxis
1. Signal pathway of a single bacterium
- the signal transfer inside the bacterium
chemoattractant stimulation -> chemoreceptor -> CheA -> CheY-P -> flagellar motor -> changed switching frequency between swimming and tumbling (biasing fraction) - the phosphrylation level of CheY can indicate the signal transferring pathway, the CheY-P concentration increases when the chemoreceptor is triggered
- the modeling result will show the optimal concentration to trigger the chemoreceptors (10-6 mol/L)
high = saturated chemoreceptors
low = cannot be detected



2. Bacterial population dynamics
An animation is made to show the movement of the population:
mainly by diffusion, biased motion with the presence of chemoattractant
based on the malate distribution in steady-state and the Spiro model
 "
"