Team:USTC-China/Human practice/The Three Laws
From 2011.igem.org

Three Laws of Synthetic Biology
1).Background: The Three Laws of Robotics, often shortened to The Three Laws or Three Laws, were written by science fiction author Isaac Asimov and later expanded upon. The rules were introduced in his 1942 short story "Runaround", although foreshadowed in earlier stories. This regulation goes like this: 1.A robot may not injure a human being or, through inaction, allow a human being to come to harm. 2. A robot must obey any orders given to it by human beings, except where such orders would conflict with the First Law. 3. A robot must protect its own existence as long as such protection does not conflict with the First or Second Law. from three laws.The Three Laws are still followed by robot scientists worldwide today, which clearly foresaw the problems that might arise between advanced robots and humans. Similarly, Synthetic Biology is faced with the same problem. If we were God, what rules should we follow so that we won't ruin the original perfect God-given world?
A. What are the problems that "Three Laws of Synthesis Biology" needs to deal with?
First, there are reports like this "A 2004 study performed near an Oregon field trial for a genetically modified variety of creeping bentgrass (Agrostis stolonifera) revealed that the transgene and its associate trait (resistant to the glyphosate herbicide) could be transmitted by wind pollination to resident plants of different Agrostis species, up to 14 km from the test field. " So "undesirable and uncontrolled gene flow into wild populations" is what we have to deal with to protect the wild types. Second, mutation is inevitable, but when mutation accumulated to a certain degree, it will generate a "Synthetic Killer Gene", that's why we need a system to inspect and then destroy the mutated ones. Last, like the Killer Bees in South America, engineering bacteria may also cause catastrophes in nature. So we responsible researchers need to set up appropriate restrictions for engineering bacteria. Based on what have been discussed above we now give our type of Three Laws:
B. 'Three Laws of Synthetic Biology'
1. Once an altered plasmid gets into bacteria that does not aim at adopting the plasmid, the suicidal mechanism of the plasmid will be executed. 2. Once the mutation accumulates to a certain degree, the suicide mechanism of the bacteria will be executed. 3. Once an infected bacterium is away from its regular surroundings, the suicide mechanism will be executed.
C. Further explanation
1. The First Law is aimed at dealing with unexpected Gene Flow between engineering bacteria and wild types. 2. The Second Law is aimed at unexpected mutations. Mutations, as we know, is inevitable and uncontrollable, thus we can now only reduce the uncertainty mutations bring about. 3. The Third Law is aimed at the possibility of biological invasion caused by escaped bacteria. Because the bacteria with changed plasmid may have much more advantages over the wild types. 4.It's easy to tell that to carry out the Three Laws we need to add more segments into the plasmid and this regulation will surely reduce the commercial value of engineering bacteria or Gene Segments, However, we believe only with great responsibility comes great power.
D. Detailed Constructions.
In order to achieve the Three Laws, several systems and methods need to be exploited. Furthermore, only if these principles be admitted by institutions and are set to be the basic rules of synthetic works worldwide can we achieve our goal of "balanced co-existence between God-given types and artificial types." That is, we need to construct new standards for synthetic biology, more parts are needed for a standard biobrick or an experiment so that the Three Laws will not be violated during all the process of construction. After some researches and discussions, we build systems that may fulfill our demands: 1.Double Plasmids Strategy (DPS): for all the synthetic works, scientists should use at least two plasmids for each artificial gene segment. (see figure 1)
a. Overview:
Plasmid A,B is created together for the aimed gene segment(the blue section).Both of them have suicidal zymogen activator sequence, suicidal zymogen sequence as well as the restrain sequence for the other. Thus, as long as A and B were together, the plasmids will work regularly. Once one of the plasmids spread into the wild, the suicidal enzyme activator will start to translate and then activate the suicidal zymogen, as a result, the plasmid would be destroyed. In this way, Law 1 would be fulfilled.
b. Further explanation:
the reason why we use two segments to start the suicidal process is that missing expression is inevitable in any cell. In order to reduce the impact of DPS towards normal cells, we add the segment of 'activator'. c. Questions: 1.What if the suicidal parts mutated? 2. What if A&B were spread into one certain cell? 3. What can be used as suicidal parts? Are there some parts that can be used for a large scale of aimed genes? 4. What would be the proper ratio between plasmid A&B?
2).Inserting New Section
a. Backdrop:
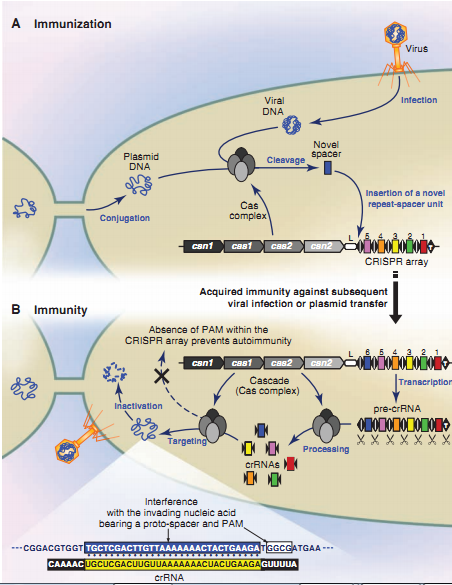
we found CRISPR/Cas System may be used as a method in DPS. CRISPR represents a series of DNA found in most archaeal (~90%) and bacterial (~40%) genomes .CRISPR, in combination with Cas proteins, forms the CRISPR/Cas system.
Overview of the CRISPR/Cas mechanism (A) Immunization process: After the insertion of exogenous DNA from viruses or plasmids, a Cas complex recognizes foreign DNA and integrates a novel repeat-spacer unit at the leader end of the CRISPR locus. (B) Immunity process: The CRISPR repeat-spacer array is transcribed into a pre-crRNA that is processed into mature crRNAs, which are subsequently used as a guide by a Cas complex to interfere with the corresponding invading nucleic acid. Repeats are represented as diamonds, spacers as rectangles, and the CRISPR leader is labeled L.
b. Modeling:
All the biobricks should be added with a CRISPR proto-spacer in the front. The proto-spacer is protected normally. Once the plasmid was spread and absorbed into any bacteria, the loci will be exposed by the restriction endonuclease, thus will be recognized by the CRISPR system and cleaned out. This model is then put together with the DPS. Plasmid 'A' contains reconstructed gene and will be added with a proto-spacer in the front, as suicidal segment, normally, will be restricted thus won't express or be recognized by the CRISPR/Cas system. However, once 'A' is spread into the wild and absorbed by the bacteria, the activator will be launched and the proto-spacer would be exposed by restriction endonuclease, thus would be destroyed by the CRISPR system.
c. Deficiency:
These viral particles have specifically mutated the proto-spacer (sequence within the invading nucleic acid that matches a CRISPR spacer), with a single mutation that allows the viruses to overcome immunity. Alternative strategies that allow viruses to escape CRISPR, such as suppressors that could interfere with crRNAs biogenesis or Cas machinery remain uncovered. d. Questions: 1.What if the proto-spacer mutates? 2. What if the CRISPR/Cas system works slower than the reconstructed plasmid and the product of the plasmid can influence the restriction endonuclease? 3.Is there any other system that we can use? 3).Bacteria: auxotroph bacteria should be used as the expectation of Law three. 4).Mutation: no good ideas so far.
Question: Is there any available systems or parts that can construct several pairs of standard plasmids as used in iGEM? 1. The Plasmids pair should be stable and be able to be used as a biobrick. And users would only need to change the working parts according to their own work. 2. The sections should have little influence to most artificial parts.
 "
"