Team:Potsdam Bioware/Labjournal/May
From 2011.igem.org
Contents |
1st Labday 12.05.2011
Preparative DNA Electrophoresis, Test Experiment 629, 654 digest
Investigators: Sascha, Jessica, Steffi, Sandrina, Nadine; introduction by Kristian, Tobias, Tim
Aim: get used to the lab
Time: 13:40
Method:
Samples digested with AscI and NheI from Tim
vector: 629 source: Tobias; prep-type: ? ; prep-date: ; clone-no: ?
insert: 654 source: Tobias; prep-type: ? ; prep-date: ; clone-no: ?
Electrophoresis
sample preparation: 30 µl 629 (digested) + 6 µl 6x loading dye solution, 30 µl 654 (digested) + 6 µl 6x loading dye
0,5 g Agarose, 50 ml TAE (1%), 2 µl GelRed (3-6µl), at 100 Volt,running time: 30 minutes
marker: GeneRuler ladder mix (Fermentas)
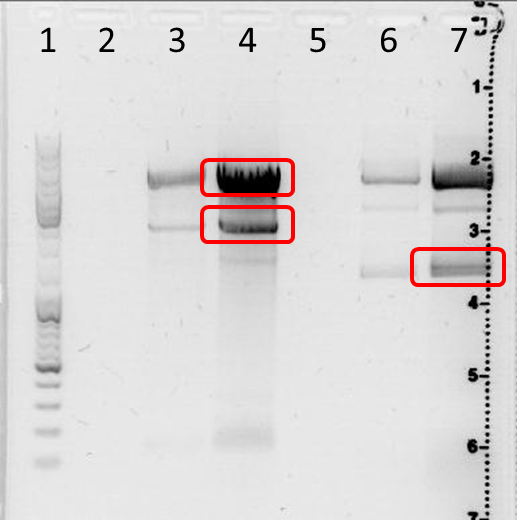
| lane | Sample | Sample/µl] | Expected size (bp) | approx. size |
|---|---|---|---|---|
| 1 |
marker |
4 µl |
||
| 3 |
629 |
5 µl | 4060, 140 | 6000, 3000, 150 |
| 4 |
629 |
31 µl | 4060, 140 | 6000, 3000, 150 |
| 6 |
654 |
5 µl | 4140, 1268 |
6000, 4000, 1700 |
| 7 |
654 |
31 µl | 4140, 1268 | 6000, 4000, 1700 (2 bands?) |
Gel Extraction
DNA excision: lane 4: 6000 bp band and 3000 bp band; lane 7: 1700 bp band
DNA purification: with Qiagen Gel Extraction Kit according to Qiagen manual (incl. isopropanol step)
Result: size estimation by eye;
lanes 3+4: plasmid was digested, small band has the expected size, two upper bands with unexpected size;
lanes 6+7: plasmid was digested; 6000 bp unexpected; 4000 bp band might correspond to expected 4140 bp band, 1700 bp band above expected 1268 bp
Conclusions:
- exact vector fragment unknown, therefore 6000 bp band and 3000 bp band were excised from lane 4, lane 7: 1700 bp band was excised;
- because of unexpected sizes marker should be compared with
another marker, plasmids should be digested to yield several fragments
for size control, maybe heat inactivation might help to resolve higher bands
Output: three plasmid fragments with 6000 bp (V1), 3000 bp (V2) and 1700 bp (I1) in 1,5 ml eppis,
stored at -20 °C, box with fragments
Ligation 629 vec frag, 654 insert
Investigators: Sascha, Jessica, Steffi, Sandrina, Nadine
Aim: create a new vector for Tim
Time: 16:30
Method:
vector: 629 source: Tobias; prep-type: ; prep-date: 12.4.2011; clone-no: ?
insert: 654 source: Tobias; prep-type: ; prep-date: 12.4.2011; clone-no: ?
Ligation: 2 µl vector, 2 µl insert, 1 µl quick ligase, 5 µl 2x quick ligase NEB, 10 min
Output: plasmid: pNoname_Tim_V1, pNoname_Tim_V2, 2 eppis labeled 'ligation', handed over to Tim
Transformation of ligation 629_vec_frag 654_insert
Investigators: Sascha, Jessica, Steffi, Sandrina, Nadine
Aim:amplification of previous ligation
Time: 16:40
Method:
ligation-samples from 12.5.11;
pNoname_Tim_V1 in BL21 (CaCl2 chemical competent, source: Tobias, date: unknown) and XL1 (CaCl2 chemical competent,
source: Tobias, date: unknown)
pNoname_Tim_V2 in BL21 (CaCl2 chemical competent, source: Tobias, date: unknown) and XL1 (CaCl2 chemical competent, source: Tobias, date: unknown)
protocol:
addition of 2 µl ligation reaction to cells in 1.5 ml Eppi,
incubation 30 min on ice,
heat shock 45 sec at 42°C,
addition of 750 µl SOC medium,
incubation at 37 °C for 45 min in Eppendorf thermomixer at 750 rpm,
centrifugation at 2000xg; for 3 min,
decandation of supernatant,
resuspension of pellet in approx. 150 µl,
plating on LB medium with 1,5 % agar, 100 µg/ml ampicillin,
storage over night at 37°C
Results and Conclusion (13.5.2011):
pNoname_Tim_V2 in BL21: 4 distinct colonies
test ligation is missing, not enough clones, pick one clone for test
digest, repeat experiment
Output four agar plates given to Tim (only one with clones)
2nd Labday 2011-05-13
Design of primers and annotation of pARW071
Investigators: Jessica, Katharina, Hanna, Nadja, Nicole
Aim: Design of primers for sequencing pARW071, annotation of pARW071
Time: 2011-05-12, 4:00 pm - 8:00 pm and 2011-05-13, 11:15 am - 12:30 am
Materials/ Software:
- Geneious Pro 5.1.7
- Oligo Calc: Oligonucleotide Properties Calculator (http://www.basic.northwestern.edu/biotools/oligocalc.html)
Method:
1. Annotation of pARW071
- Alignment of pARW071 and pDrive_Qiagen (PCR cloning vector)
- Alignment of pARW071 and AM943877 (Microcystis aeruginosa NIES-298 microviridin biosynthetic gene cluster, strain NIES-298)
2. Primer design
- melting temperature based on 4+2 method, sum has to be 56°C
- check specific binding using dotplot feature (parameters: low sensitivity: 10, tick at reverse complementary) of Genious Pro 5.7.1
- Check self-complementarity and melting temperature using Oligo Calc
Results:
Primer sequences
- sf_bb_1_1 ACAATTCACTGGCCGTCG
- sf_bb_2_1 AAGATCCTTGAGAGTTTTCG
- sf_bb_3_1_n AAATTCCAACATGGATGCTG
- sf_bb_4_1 TAATACGACTCACTATAGGG
- sf_mdn_1_1 ACCAACAAATACAGAGCCAG
- sf_mdna_1 GGTTATTGCAGGTGGTTAG
- sf_mdnc_1 ATCGCTTCCCTACAGAGG
- sf_mdnd_1 CTCCTAACCCCACC
- sf_mdne_1_n AAGGTTTTTGGGTCTTTATCG
- sf_mdne_2_1 CGATAAATTCTCCACAATTACC
- sf_mdne_3_1 AATTGAGAAAAAGCAAATAACC
- sf_mndb_1 GTTATTACAACTTCGTTTGG
pARW071 with primers and annotations
Conclusion:
Primer ordering by Sigma-Aldrich
Further tasks
- Midi Prep of E. coli carrying pARW071 and pARW089
- Sequencing of pARW071
3rd Labday 2011-05-23
Overnight cultures of pARW071 and pARW089 carrying cells
Investigators: Jessica, Nadja, Nicole
Time: 2011-05-23, 5:30 pm - 7:30 pm
Materials:
- LB medium
- Ampicillin 100 µg/ ml
Clones:
1. pARW071 pDrive-mdnA-E, N843(Top10 cells) - clone 1; Source: Elke Dittmann; Date of plate:
2. pARW089 pDrive-mdn cluster with Aat II/ Ehe I restriction sites before and afterwards the mdnA gene (XL1O gold cells) - clone 1; Source: Elke Dittmann; Date of plate:
Method:
Inoculation of one clone each in 100 ml LB medium containing ampicillin
shaking over night at 37°C, 300 rpm, approx. 19 hours
Further tasks:
- Midiprep
- Sequencing of complete pARW071
- Sequencing of mdnA in pARW089
Output:2 over night cultures pARW071 in Top10 cells, pARW089 in XL10 cells
4th Labday 2011-05-24
Midiprep of pARW071 and pARW089 carrying cells
Investigators: Nadja, Nicole
Aim:
- Midiprep of pARW071 and pARW089 carrying cells
- Glycerol stocks of E. coli cells carrying pARW071 and pARW089
Time: 2011-05-24, 3:00 pm - 7:00 pm
Materials:
- Qiagen Plasmid Plus Midi Kit
- Glycerol
- over night cultures of pARW089 and pARW071 carrying cells
Method:
1. Glycerol stocks
- 700 µl of cell suspension
- 300 µl glycerol
- gently mixed and frozen by -80°C
2. Midiprep
- using Qiagen Plasmid Plus Midi Kit Quick-Start Protocol, Media: UP_QIAGEN_Plasmid_Plus_Midi_Kit.pdf
Output: DNA Eppi Tube labelled with "1" for pARW071 and "2" for pARW089, storage in pink DNA box
Further tasks:
- DNA concentration determination
- Sequencing of complete pARW071
- Sequencing of mdnA in pARW089
5th Labday 2011-05-25
Overnight culture of E.coli cells carrying MRC and N298 fosmids
Investigators: Nadja, Nicole
Time: 2011-05-25, 4:00 pm - 7:00 pm
Materials:
- LB medium
- 12.5 µg/ml Chloramphenicol
Method:
One clone in approx. 2 ml LB medium
- N298 fosmid (mdnB) Epi cells - plate/clone 1
- N298 fosmid (mdnB) Epi cells - plate/clone 2
- MRC fosmid (mdnJ variants) Epi cells - clone 1
- MRC fosmid (mdnJ variants) Epi cells - clone 1
shaking over night at 37°C, 300 rpm
Output:
- over night cultures
Further tasks:
- Glycerol stocks of each clone
6th Labday 2011-05-26
Glycerol stocks of E.coli cells carrying MRC and N298 fosmids
Investigators: Nadja, Nicole
Aim: Generating stocks for further research
Time: 2011-05-26,17:45-18:15
Materials:
- Glycerol
- N298 fosmid (mdnB) Epi cells - plate/clone 1
- MRC fosmid (mdnJ variants) Epi cells - clone 1
Method:
- 700 µl of cell suspension
- 300 µl glycerol
- mixed gently and frozen by -80°C
7th Labday 2011-05-27
Preparation of samples for sequencing
Investigators: Jessica, Nicole, Nadja
Aim: Prepare mixtures for sequencing
Time: 2011-05-27,13:45-15:45
Materials:
- Nanodrop
- Primers ordered previously by Sigma Aldrich --> see labday 2
- Isolated DNA of pARW071 and pARW089
Method:
1. Preparation of primer stocks
- Primers arrived dry
- Adding ddH2O (based on manufacture's sheet)
- Reached concentration 100 µM
2. DNA concentration determination
- Using nanodrop
3. Preparing sequencing mixtures
a. Dilution of DNA and primers
- Primers: needed concentration = 10 pmol/ µl, volume needed: 20 µl
- DNA: needed concentration = 60 ng/ µl, volume needed: 20 µl per 8 sequencing reactions
b. Preparation of sequencing tubes for sequencing by GATC
- For each tube one barcode, green = DNA, yellow = primer
- 2 tubes with DNA (pARW071 and pARW089)
- 12 tubes with primers (sf_mdna_1 is used for pARW071 and pARW089)
Results:
1. Concentration of plasmid DNA
- pARW071: 626.7 ng/ µl
- pARW089: 857.3 ng/ µl
2. Sequencing ordered by GATC
Further tasks:
- Analysis of sequencing of pARW071 and pARW089 (hopefully the results will arrive on monday)
- Testing PCR of the complete pARW071 and pARW089, respectively
- Inserting myc-tag
- Cloning mdnA in phagemid
- Cloning gene 3 in pARW089
8th Labday 2011-05-30
Searching for promotor region and primer design
Investigators: Jessica, Nicole, Sabine, Sandrina, Steffi, Nadja
Aim: Find the putative mdn promoters and design a primer (r_bb_1) which will be used for a PCR of the whole mdn
Time: 2011-05-30,15:00-17:50
Materials:
- Geneious Pro 5.1.7
- Oligo Calc: Oligonucleotide Properties Calculator (http://www.basic.northwestern.edu/biotools/oligocalc.html)
- Some online tools to search for the promotors:
1. http://linux1.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfindb
2. http://bioinformatics.biol.rug.nl/websoftware/ppp/ppp_start.php
3. http://genie.dartmouth.edu/scope/results.php?jobid=111290
Method:
1. Primer design
- melting temperature based on 4+2 method, sum has to be 56°C
- check specific binding using dotplot feature (parameters: low sensitivity: 10, tick at reverse complementary) of Genious Pro 5.7.1
- Check self-complementarity and melting temperature using Oligo Calc
2. Promoter search
- Application of software tools
- Annotation of pARW071
Results:
1. Primer sequence
- r_bb_1 CGACGAATTCAGATTAGGAA
2. Search for the promotors
- Search results see pdf file Media: UP_2011-05-30_PromoterSearchResults.pdf?
Further tasks:
- PCR of mdn
- Talk with Elke about the promoter
9th Labday 2011-05-31
Analysis of sequencing results and Primer design
Investigators: Jessica, Nicole, Sandrina, Nadja
Aim: Confirm pARW071 sequence and order primer (r_mdna_1) for PCR of mdnA of pARW089
Time: 2011-05-31,11:45-16:00
Materials:
- Geneious Pro 5.1.7
- Oligo Calc: Oligonucleotide Properties Calculator (http://www.basic.northwestern.edu/biotools/oligocalc.html)
Method:
1. Primer design
- melting temperature based on 4+2 method, sum has to be 56°C
- check specific binding using dotplot feature (parameters: low sensitivity: 10, tick at reverse complementary) of Genious Pro 5.7.1
- Check self-complementarity and melting temperature using Oligo Calc
2. Sequencing analysis
- Alignment of sequencing results with pARW071
Results:
1. Primer sequence
- r_mdna_1 CGGTGTAATCAAGAAAAGT
2. Sequencing analysis
- pARW071
| Primer | Mismatch | Consensus location |
|---|---|---|
| sf_bb_1 1 | - | - |
| sf_bb_2 1 | - | - |
| sf_bb_3 1n | - | - |
| sf_bb_4 1 | 3 | 677, 777, 857 |
| sf_mdna_1 | - | - |
| sf_mdnb_1 | / | / |
| sf_mdnc_1 | 3 | 2678, 2860, 2960 |
| sf_mdn | 2 | 3780, 4270 |
| sf_mdne_1n | 3 | 4718, 5041, 5158 |
| sf_mdne_2 1 | 3 | 5480, 5800, 6050 |
| sf_mdne_3 1 | 1 | 6824 |
- pARW089
| Primer | Mismatch | Consensus location |
|---|---|---|
| sf_mdna_1 | - | - |
- Legend: - = no mismatches; / = sequencing did not work
Further tasks:
- Check how serious the consequences of the found mismatches are
 "
"