Team:Freiburg/Notebook/14 July
From 2011.igem.org

Contents |
red light receptor
3A-assembly: Digest
Investigators: Jakob
3A-assembly of S22 (ho1)+S21a (terminator) and S23 (pcyA)+S21a (terminator)
Digestion
| Name:Jakob | Date: 14.07.2011 |
| Project Name: 3A-assembly of pcyA+ terminator, ho1+terminator | |
Procedure
1. add H2O (16μl-DNA )
2. 2μl NEB4 buffer
3. 2 μl 10x BSA (used 1:10 diluted )
4. DNA (500 ng) (Vector:Insert ratio 1:3 in following Ligation)
5. 1 μl restriction enzymes
6. heat for 1-2 hours 37°C (6 hours if time)
7. heat for 20 minutes 80°C (inactivation of enzymes)
8. keep at 4°C if you cannot continue
Measured DNA-concentration with Nanodrop to calculate the volume of DNA to do the digestion:
| Sample Name | DNA concentration (μg/μl) |
| Vector pSB1T3 | 20 |
| terminator | 38.8 |
| ho1 | 176 |
| pcyA | 173 |
Restriction enzymes you need to cut the vector, insert1 and insert 2:
| Components | | | |
| DNA (μl) | 2 | Ho1 2.5
pcyA 2.5 | Terminator 5.5
terminator 5.5 |
| BSA (10x) (2μl) | |||
| NEB4 Buffer (2μl) | |||
| Enzyme 1 (1μl) | EcoRI | EcoRI | XbaI |
| Enzyme 2 (1μl) | PstI | SpeI | PstI |
| H2O (14 μl- DNA) | |||
| In total 20 μl | |||
3A-assembly: Ligation and transformation
Investigators: Julia
Ligation of S22 (ho1)+S21a (terminator) and S23 (pcyA)+S21a (terminator) in vector pSB1T3.
L23: Ligation of S23+S21a in pSB1T3 L22: Ligation of S22+S21a in pSB1T3 Ligation
| Name:Julia | Date:14.07 |
| Continue from Date14.07 Name
Experiment 3A-assembly | |
| Project Name:3A-assembly of pcyA+ terminator, ho1+terminator | |
Procedure
total volume 20 μl
1. add H2O(17 μl -X-Y-Z)
2. add 2 μl Ligase Buffer 10x
3. add Insert 1, Insert 2 (when proceeding from 3A digestion use 2 μl of each)
4. add Vector (20ng needed. When proceeding from 3A digestion use 2 μl)
5. Add 1 μl T4-DNA Ligase
6. Incubate 10-30 min at room temperature
7. heat for 20 minutes at 80°C
8. store at -20°C or directly proceed to transformation
| Name of part | Ratio Insert:Vector
1:1 | Volume (μl) | |
| X insert 1 | ho1/ pcyA | 2 | |
| Y insert 2 | terminator | 2 | |
| Z vector | pSB1C3 | 2.5 | |
| H2O | 11 |
Competent neb10 strain was transformed with 2μl ligation reaction and plated out.
PCR
Investigators: Julia
PCR of the red light receptor, cph8.
Primer:
| P10 | atatgaattcgcggccgcttctagATGGCCACCACCGTAC |
| P11 | atatatgcatctgcagcggccgctactagtaTTACCCTTCTTTTGTCaTGCC |
DNA template:
- BBa_I150010
Programm:
2min 55/60
Lysis cassette
Quickchange PCR
| Name: Theo
| Date: 14.07.2011 |
| Continue from Experiment Lysis Cassette
| |
| Project Name: Correct number of nucleotides between RBS and ATG of temp. sensitive repressor from
Lysis Device | |
PCR-Mixture for one Reaction:
For a 50 µl reaction use
| 32,5µl | H20 | Name |
| 10µl | 6x HF Phusion Buffer | |
| 2.5µl | Primer fw | P29 (1:10) |
| 2.5µl | Primer dw | P30 (1:10) |
| 1µl | dNTPs | |
| 1µl | DNA-Template | M15a (from S15->diluted to 5ng/µl) |
| 0.5 µl | Phusion (add in the end) |
What program do you use?
| | |||
| | | | |
| | |||
| |
Precipitator
Test-digest
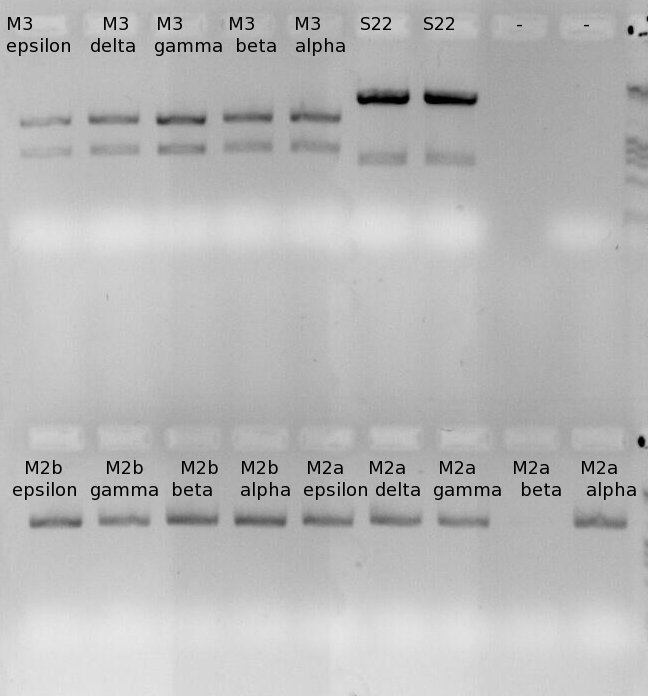
Investigators: Rüdiger
Testdigest of the minipreps, Sophie did (june, 13).
- Enzyme 1: PstI
- Enzyme 2: EcoRI
Image of different samples from miniprep.
 "
"
 Contact
Contact