Team:Washington/Parts
From 2011.igem.org
(Difference between revisions)
(→Data for Favorite New Parts) |
(→Improved Parts) |
||
| (85 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
<center><big><big><big><big>'''Data Page'''</big></big></big></big></center><br><br> | <center><big><big><big><big>'''Data Page'''</big></big></big></big></center><br><br> | ||
| - | <center>< | + | <center><big>An overview of the 2011 UW iGEM team's summer projects</big></center> |
| - | Image:Diesel | + | [[File:Washington_Spacer.jpg|1px]] |
| - | Image:Gluten | + | [[Image:UW Diesel Front Page.png|300px|link=https://2011.igem.org/Team:Washington/Alkanes/Background]] |
| - | Image: | + | [[File:Washington_Spacer.jpg|20px]] |
| - | + | [[Image:UW Gluten Front Page.png|300px|link=https://2011.igem.org/Team:Washington/Celiacs/Background]] | |
| - | + | [[File:Washington_Spacer.jpg|20px]] | |
| - | + | [[Image:UW Toolkits Front Page.png|300px|link=https://2011.igem.org/Team:Washington/Magnetosomes/Background]] | |
| + | [[File:Washington_Spacer.jpg|5px]] | ||
| + | {| style="background: white; text-align: center; width: 970px;" align="center" border="0" | ||
| + | | '''Make It: Diesel Production''' | ||
| + | | '''Break It: Gluten Destruction''' | ||
| + | | '''iGEM Toolkits: Gibson Assembly<br>and Magnetosomes''' | ||
| + | |} | ||
| + | {| style="background: white; text-align: left; width: 965px;" border="0" | ||
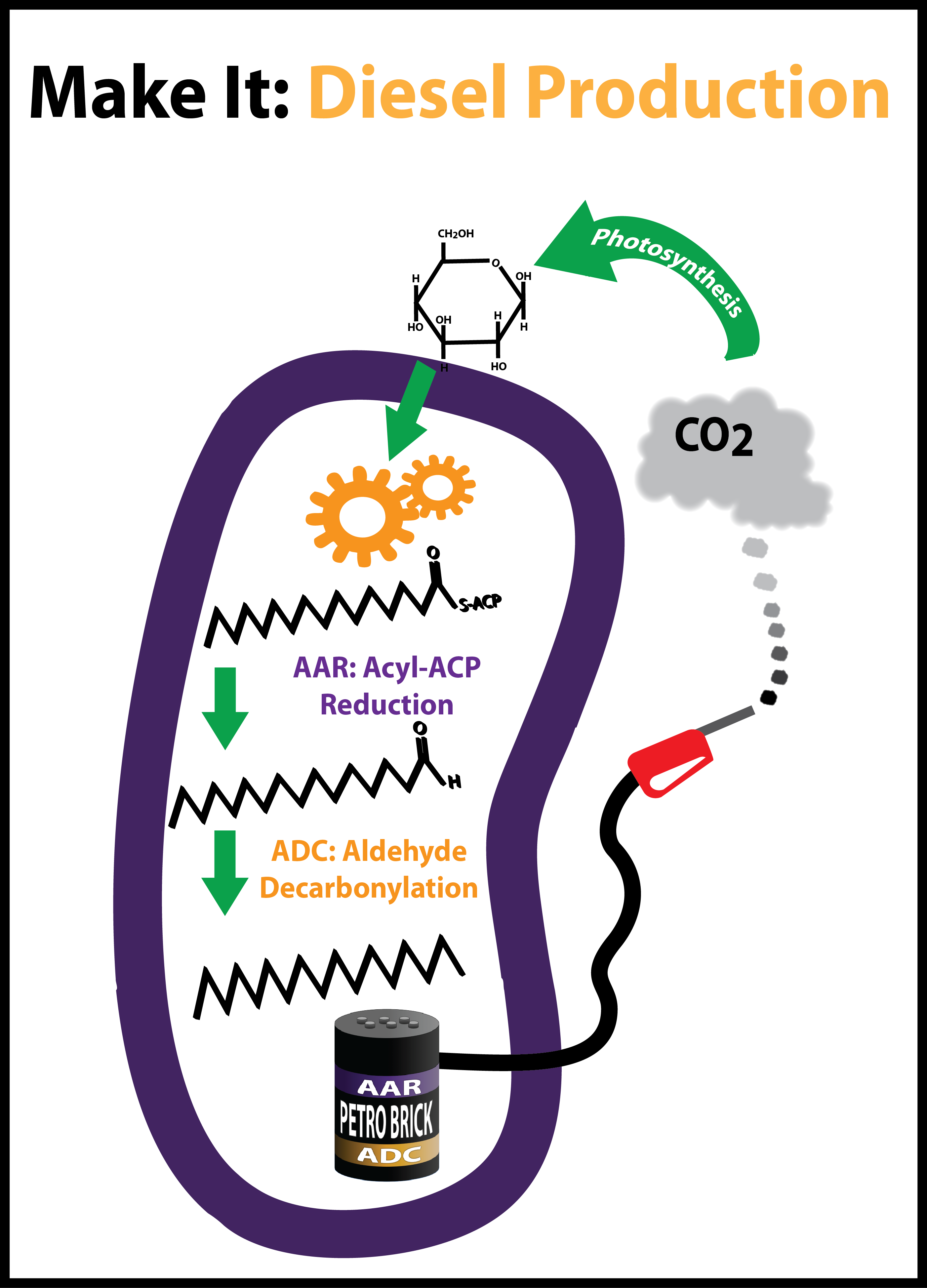
| + | |We designed and constructed a modular alkane production BioBrick, the PetroBrick, to generate alkanes, the main constituent of diesel. By expressing this BioBrick in ''E. coli'', we were able to produce alkanes of yields over 100 mg/mL. | ||
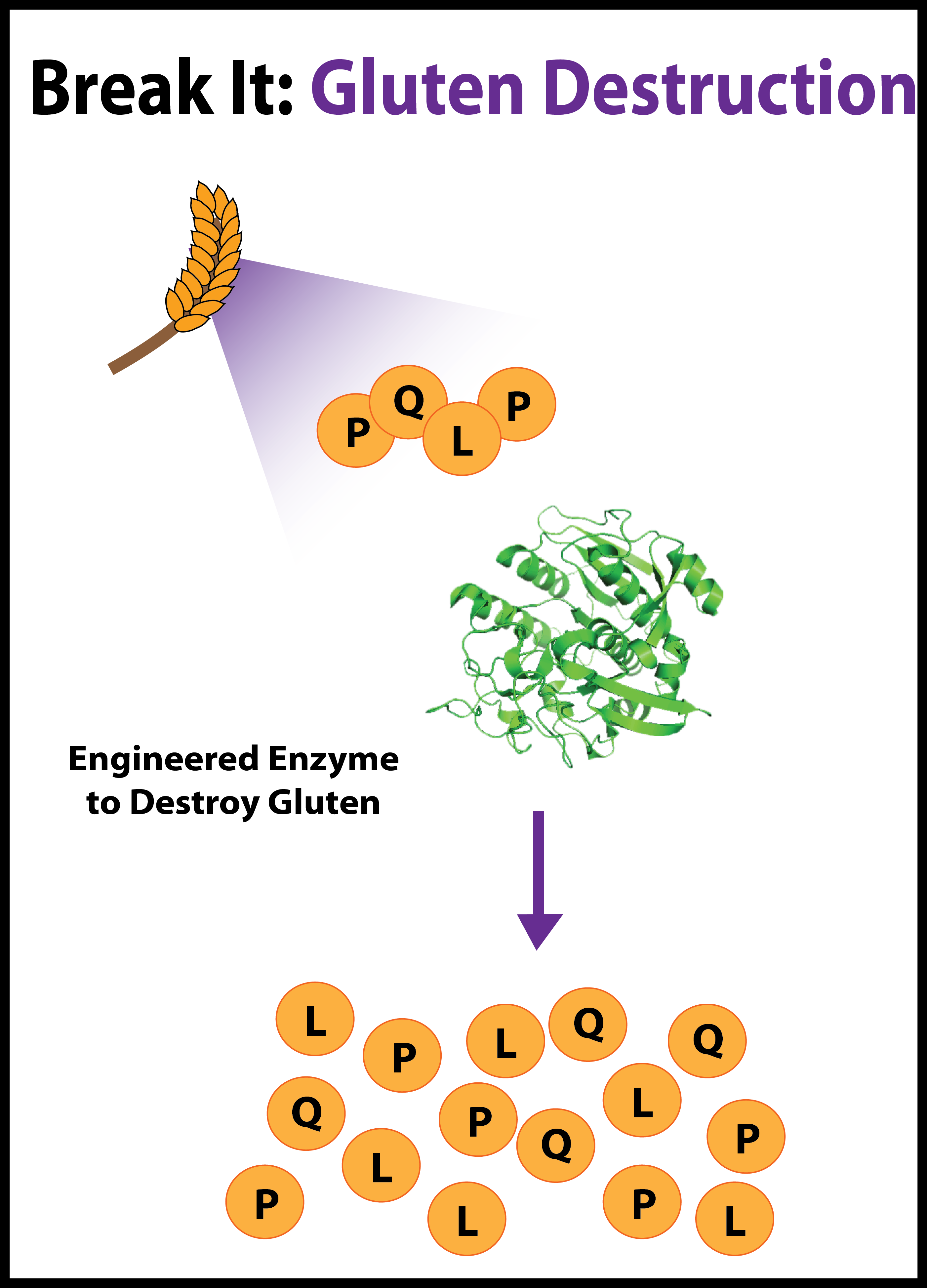
| + | |Gluten intolerance stems from an inappropriate immune response to PQLP, the most common motif in the immunogenic peptide. We reengineered a protease, active at low pH, for strongly enhanced activity against PQLP. | ||
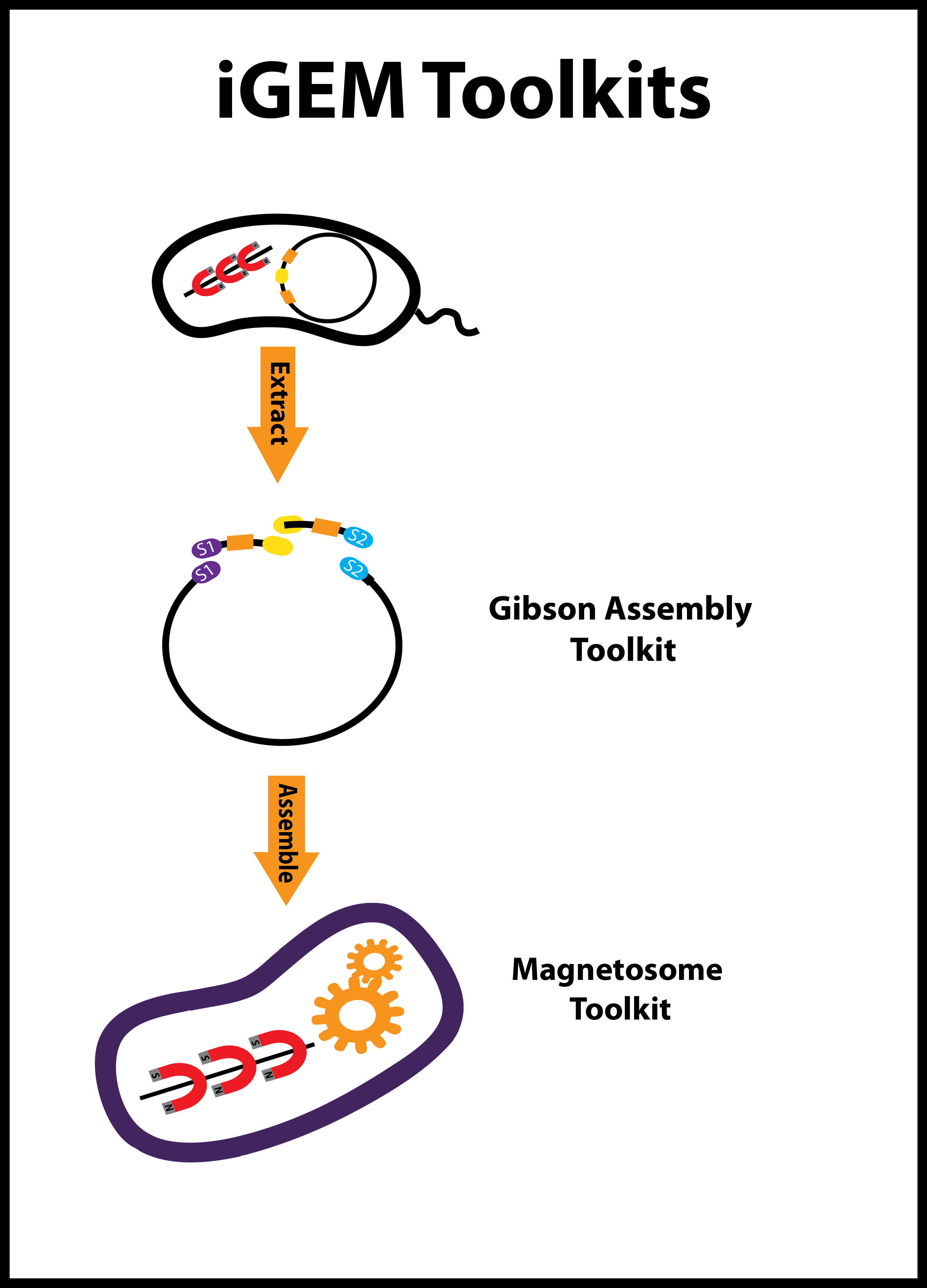
| + | |We built two iGEM toolkits. The first is a set of five BioBrick vectors optimized for Gibson assembly. The second is a set of genes essential for magnetosome formation characterized in ''E. coli''. | ||
| + | |} | ||
='''Data Summary'''= | ='''Data Summary'''= | ||
| - | ==Data for Favorite New Parts== | + | ==''Data for Favorite New Parts''== |
==='''Diesel Production'''=== | ==='''Diesel Production'''=== | ||
| - | [http://partsregistry.org/Part:BBa_K590025 BBa_K590025 ''' | + | :: '''1, 2.''' [http://partsregistry.org/Part:BBa_K590025 BBa_K590025: '''The PetroBrick'''] - A modular and open platform for the biological production of diesel fuel. The PetroBrick consists of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590032 AAR] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590031 ADC], each behind a standard Elowitz RBS. All of this is under regulation by a high constitutive promoter in pSB1C3. |
| - | + | :: '''3''' [http://partsregistry.org/Part:BBa_K590064 '''The FabBrick'''] - An add-on module to the PetroBRick that causes the production of odd chain length Fatty acids. This part consists of [ http://partsregistry.org/Part:BBa_K590034 FabH2] expressed on a [http://partsregistry.org/wiki/index.php?title=Part:BBa_K314103 low copy number IPTC inducible vector] These are converted into even chain length alkanes by [http://partsregistry.org/Part:BBa_K590025 BBa_K590025 the PetroBrick], completing the spectrum of linear alkane compounds that can be produced using the | |
| - | A modular and open platform for the biological production of diesel fuel. The | + | |
| - | + | ||
| - | [http://partsregistry.org/ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | [http://partsregistry.org/ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | [http://partsregistry.org/wiki/index.php?title=Part: | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | [http://partsregistry.org/ | + | |
| - | + | ||
| - | + | ||
==='''Gluten Destruction'''=== | ==='''Gluten Destruction'''=== | ||
| - | [http://partsregistry.org/wiki/index.php?title=Part: | + | :: '''4.''' [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590087 BBa_K590087: '''KumaMax''']- A modified version of the enzyme Kumamolisin, a protease ofthe sedolisin family native to ''Alicyclobacillus sendaiensis'' known to be active at low pH and elevated temperatures. To Kumamolisin, the mutations N291D, G319S, D358G, D368H increase activity to the PQLP peptide, an antigenic epitope in gliadin, 118-fold. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==='''Gibson Assembly Toolkit'''=== | ==='''Gibson Assembly Toolkit'''=== | ||
| - | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590010 BBa_K590010 ''' | + | ::'''5.''' [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590010 BBa_K590010: '''pGA1A3'''], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590011 BBa_K590011: '''pGA1C3'''], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590012 BBa_K590012: '''pGA4C5'''], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590013 BBa_K590013: '''pGA4A5'''], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590014 BBa_K590014: '''pGA3K3'''] - These are plasmid backbones based on the bglBrick standard (BBF RFC 21) and optimized for use in Gibson assembly. These vectors are suitable replacements for the equivalent pSB vectors for iGEM teams using Gibson cloning to assemble their constructs. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590011 BBa_K590011 ''' | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590012 BBa_K590012 ''' | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590013 BBa_K590013 ''' | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590014 BBa_K590014 ''' | + | |
| - | + | ||
| - | + | ||
==='''Magnetosome Toolkit'''=== | ==='''Magnetosome Toolkit'''=== | ||
| - | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590015 BBa_K590015 '''sfGFP_mamK_pGA1C3'''] | + | :: '''6.''' [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590015 BBa_K590015: '''sfGFP_mamK_pGA1C3'''] - This part consists of the ''mamK'' gene from ''Magnetospirillum magneticum'' strain AMB-1, as a superfolder GFP fusion, in the pGA1C3 backbone. MamK creates an actin-like filament that orients itself along the long-axis of the cell and acts as the scaffold for the alignment of magnetosome vesicles. |
| - | + | ||
| - | This part consists of | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | :: '''7.''' [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590016 BBa_K590016 '''sfGFP_mamI_pGA1C3'''] - This part consists of ''mamI'' gene from ''Magnetospirillum magneticum'' strain AMB-1, as a superfolder GFP fusion, in the pGA1C3 backbone. MamI is a membrane-localized protein required for magnetosome vesicle formation that also binds the poly-MamK filament. | |
| - | + | ||
| - | + | ||
| - | + | ||
| + | ==''Data for Existing Parts''== | ||
| - | + | ::'''8.''' [http://partsregistry.org/Part:BBa_K314100:Experience K314100: '''High Constitutive Expression Cassette'''] (Washington, iGEM 2010) - We used this part to express our Petrobrick, found that it works well for expression, and entered this information in the part experience page. | |
| - | + | ::'''9.''' [http://partsregistry.org/Part:pSB1A3:Experience pSB1A3] - As part of the Gibson Vector Toolkit we characterized the cloning efficiency of this plasmid backbone for Gibson assembly, using the prefix and suffix regions as primers. We found that pSB1A3 had a proper insert efficiency of 11%, compared to 99% for the equivalent Gibson-optimized pGA1A3 vector. | |
| - | + | ||
| + | ==''Improved Parts''== | ||
| + | ::'''10.''' [http://partsregistry.org/Part:BBa_K590059 BBa_K590059], [http://partsregistry.org/Part:BBa_K590060 BBa_K590060], [http://partsregistry.org/Part:BBa_K590061 BBa_K590061: '''LuxC, D, and E'''] (Cambridge, iGEM 2010) - Formerly part of the [http://partsregistry.org/Part:BBa_K325909 LuxBrick] the genes ''luxC, D,'' and ''E'' were not separated or codon-optimized. We codon-optimized these genes and put them under the control of standard Elowitz RBS's (B0034). This was accomplished by the Alternate Aldehyde branch of the Alkane Production team. | ||
='''All Submitted Parts'''= | ='''All Submitted Parts'''= | ||
<center><groupparts>iGEM011 Washington</groupparts></center> | <center><groupparts>iGEM011 Washington</groupparts></center> | ||
Latest revision as of 18:09, 26 October 2011
| Make It: Diesel Production | Break It: Gluten Destruction | iGEM Toolkits: Gibson Assembly and Magnetosomes |
| We designed and constructed a modular alkane production BioBrick, the PetroBrick, to generate alkanes, the main constituent of diesel. By expressing this BioBrick in E. coli, we were able to produce alkanes of yields over 100 mg/mL. | Gluten intolerance stems from an inappropriate immune response to PQLP, the most common motif in the immunogenic peptide. We reengineered a protease, active at low pH, for strongly enhanced activity against PQLP. | We built two iGEM toolkits. The first is a set of five BioBrick vectors optimized for Gibson assembly. The second is a set of genes essential for magnetosome formation characterized in E. coli. |
Data Summary
Data for Favorite New Parts
Diesel Production
- 1, 2. BBa_K590025: The PetroBrick - A modular and open platform for the biological production of diesel fuel. The PetroBrick consists of AAR and ADC, each behind a standard Elowitz RBS. All of this is under regulation by a high constitutive promoter in pSB1C3.
- 3 The FabBrick - An add-on module to the PetroBRick that causes the production of odd chain length Fatty acids. This part consists of [ http://partsregistry.org/Part:BBa_K590034 FabH2] expressed on a low copy number IPTC inducible vector These are converted into even chain length alkanes by BBa_K590025 the PetroBrick, completing the spectrum of linear alkane compounds that can be produced using the
Gluten Destruction
- 4. BBa_K590087: KumaMax- A modified version of the enzyme Kumamolisin, a protease ofthe sedolisin family native to Alicyclobacillus sendaiensis known to be active at low pH and elevated temperatures. To Kumamolisin, the mutations N291D, G319S, D358G, D368H increase activity to the PQLP peptide, an antigenic epitope in gliadin, 118-fold.
Gibson Assembly Toolkit
- 5. BBa_K590010: pGA1A3, BBa_K590011: pGA1C3, BBa_K590012: pGA4C5, BBa_K590013: pGA4A5, BBa_K590014: pGA3K3 - These are plasmid backbones based on the bglBrick standard (BBF RFC 21) and optimized for use in Gibson assembly. These vectors are suitable replacements for the equivalent pSB vectors for iGEM teams using Gibson cloning to assemble their constructs.
Magnetosome Toolkit
- 6. BBa_K590015: sfGFP_mamK_pGA1C3 - This part consists of the mamK gene from Magnetospirillum magneticum strain AMB-1, as a superfolder GFP fusion, in the pGA1C3 backbone. MamK creates an actin-like filament that orients itself along the long-axis of the cell and acts as the scaffold for the alignment of magnetosome vesicles.
- 7. BBa_K590016 sfGFP_mamI_pGA1C3 - This part consists of mamI gene from Magnetospirillum magneticum strain AMB-1, as a superfolder GFP fusion, in the pGA1C3 backbone. MamI is a membrane-localized protein required for magnetosome vesicle formation that also binds the poly-MamK filament.
Data for Existing Parts
- 8. K314100: High Constitutive Expression Cassette (Washington, iGEM 2010) - We used this part to express our Petrobrick, found that it works well for expression, and entered this information in the part experience page.
- 9. pSB1A3 - As part of the Gibson Vector Toolkit we characterized the cloning efficiency of this plasmid backbone for Gibson assembly, using the prefix and suffix regions as primers. We found that pSB1A3 had a proper insert efficiency of 11%, compared to 99% for the equivalent Gibson-optimized pGA1A3 vector.
Improved Parts
- 10. BBa_K590059, BBa_K590060, BBa_K590061: LuxC, D, and E (Cambridge, iGEM 2010) - Formerly part of the LuxBrick the genes luxC, D, and E were not separated or codon-optimized. We codon-optimized these genes and put them under the control of standard Elowitz RBS's (B0034). This was accomplished by the Alternate Aldehyde branch of the Alkane Production team.
 "
"