Team:Washington/Magnetosomes/Magnet Toolkit
From 2011.igem.org
| Line 218: | Line 218: | ||
|} | |} | ||
| - | Using our two genes of interest, we | + | Using our two genes of interest, we created C-terminal sfGFP fusions so we could track the localization of each gene separately within ''E.coli.'' |
{| border="1" | {| border="1" | ||
| Line 234: | Line 234: | ||
|} | |} | ||
| - | The results we obtained with our sfGFP fusions inside ''E.coli'' | + | The results we obtained with our sfGFP fusions inside ''E.coli'' were comparable to those done through other studies in the host organism ''Magnetospirillum magneticum''. In both images |
Revision as of 19:55, 21 September 2011
Magnetosome Toolkit
What are magnetosomes? Where do they come from?
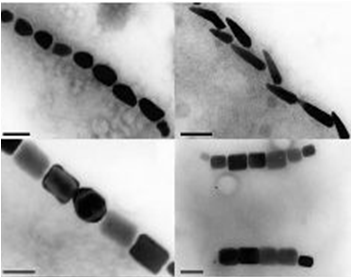
Magnetotactic Bacteria are prokaryotic organisms which possess the unique ability to align themselves along a magnetic field. This form of taxis is made possible by the formation of a magnetosome formation. Magnetosomes are small invaginations of the bacterial cell membrane that contain magnetite particles
These particles range in size between 20 and several hundred nanometers and are aligned in one or several chains along the long axis of the bacteria. These particles act together to form a magnetic dipole across the bacteria, allowing it to perceive the earth’s magnetic field. Magnetotactic bacteria are microaerophilic; therefore, the magnetosome is currently thought to help aid the organism in its search for the perfect oxygen level from a three dimensional space (in all directions) to a one dimensional space along a single path.
A Closer look at Magnetosome Formation
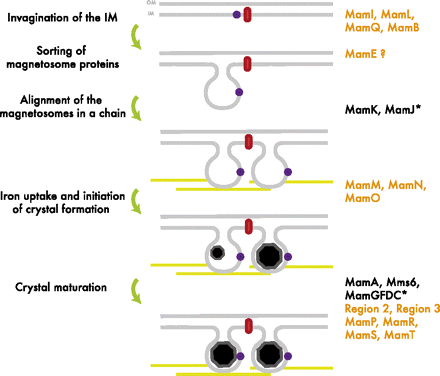
The formation of the magnetosome organelle is a highly regulated, step-wise process requiring a cascade of essential genes. The process is generally hypothesized as four stages: i) membrane invagination, ii) acquiring minerals for magnetite formation, iii) iron-oxidation and reduction, iv) magnetite nucleation and morphology regulation. Earlier gene products must be present for later gene products to be formed as shown in the diagram below: [http://www.pnas.org/content/107/12/5593.full.pdf+html]:
What did the UW iGEM team do with Magnetotactic Bacteria?
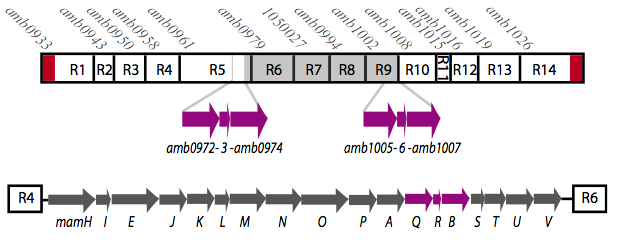
It is thought that many of the essential genes associated with magnetosome formation are located within a well-conserved region known as the magnetosome island (MAI). The MAI consists of 14 gene clusters labeled R1-R14 (see diagram below).Our team focused on the genes of the mamAB gene cluster (R5) as they were previously shown to be essential for magnetosome membrane biogenesis in AMB-1 (diagram show below).[http://www.pnas.org/content/107/12/5593/F1.expansion.html].
The goal of our project was to extract all the essential genes from (R5) required for magnetosome formation and express them in E.coli. This was done in order to understand more about magnetosome formation and the magnet synthesis mechanism because many of the genes' functions are still unknown in the host species. Using the information we have gained, we have organized a Magnetosome Toolkit containing most of the essential genes for proper magnetosome formation. Ultimately, we would like to continue expanding the magnetosome toolkit to have enough parts to show complete magnetosome formation within E.coli.
About the Magnetosome Toolkit:
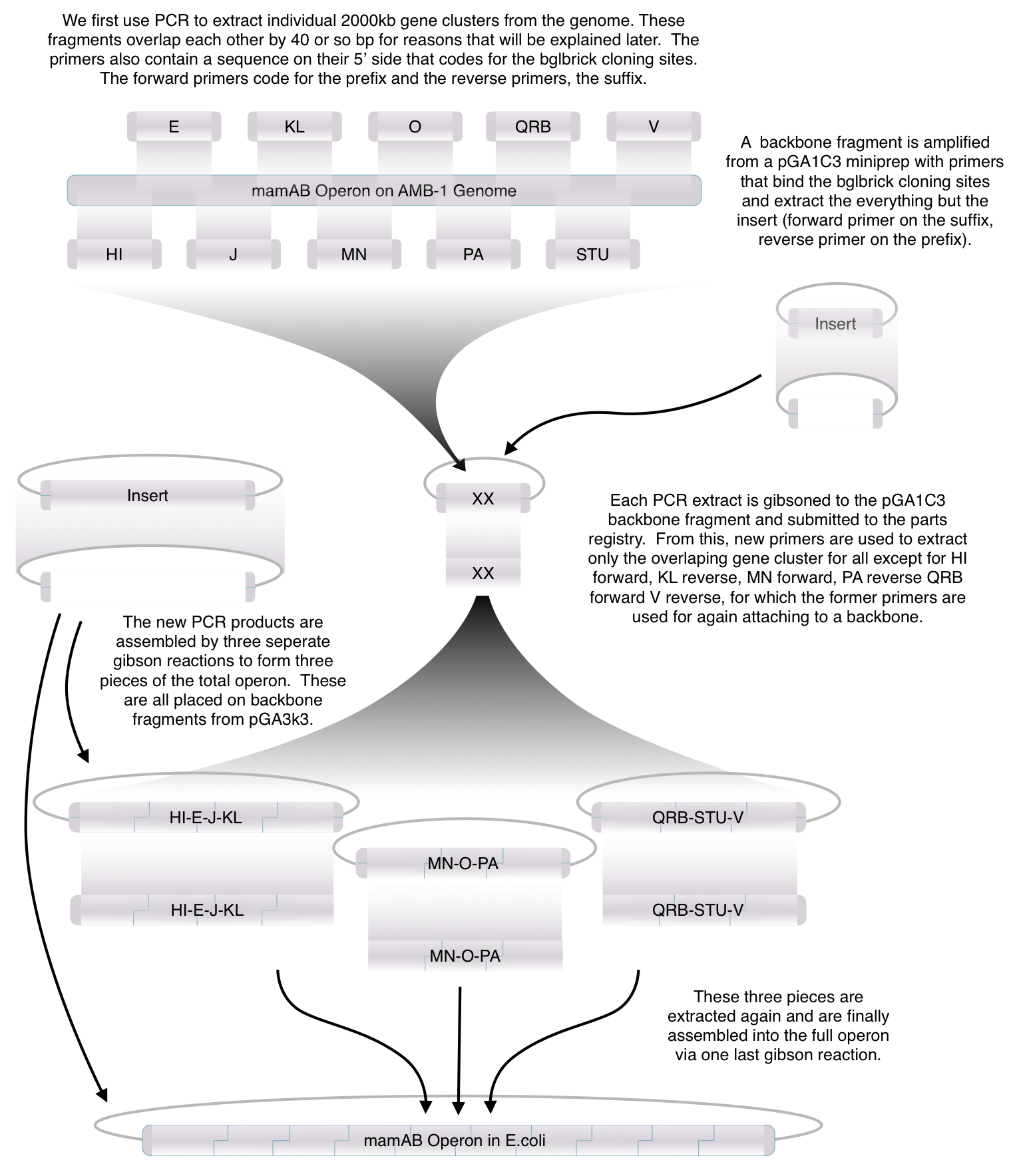
Using standard synthetic biology protocols and the vectors we created in our Gibson Assembly Toolkit, our team was able to create a "Magnetosome Toolkit" consisting of the most basic parts required for magnetosome formation. Providing this toolkit will help allow future iGem teams to manipulate and further understand magnetosome formation to eventually synthesize magnets in various types of bacteria.
Toolkit construction and mamAB assembly in E.coli
Before piecing together the 16 kb genome of the mamAB gene cluster within the magnetosome island (MAI), we extracted out the genes in the following groups:
| Gene groups | Length (bp) |
|---|---|
| mamHI | 1541 |
| mamE | 2172 |
| mamJ | 1538 |
| mamKL | 1336 |
| mamMN | 2323 |
| mamO | 1914 |
| mamPA | 1493 |
| mamQRB | 2029 |
| mamSTU | 2030 |
| mamV | 1002 |
Using standard protocols and our high-copy pGA vectors, these genes were extracted from the host genome and characterized to confirm their accuracy.
As previously noted, magnetosome formation within the host-organism, Magnetospirillium magneticum, strain AMB-1, is a highly regulated step-wise process. As shown in Fig. 2, some genes encode for an invagination in the inner membrane, other genes which help align the magnetosomes into their characteristics chains, and others which regulate the biomineralization of magnetic particles. Our team chose to focus on genes specifically related to magnetosome scaffolding/alignment since they are the essential foundation for magnetosome development. In addition, the creation of a scaffold to which other genes localize is highly applicable to systems in synthetic biology. (for more information, please see our Future Directions page)
Our genes of interest were mamK and mamI as they have functions related to localization of the magnetosome. Specifically, mamK is a bacterial actin-like cytoskeleton protein required for proper alignment of the magnetosomes in a chain. mamK is also shown to localize the mamI, which is loss inhibits membrane formation. (for other gene functions, see the table below):
| Gene | AMB Number | Cluster Membership | Member of 28 genes list? (specific*/related**) | Function Summary (Vesicle chain formation, and/or biomineralization) | Gene Function |
|---|---|---|---|---|---|
| mamH | amb0961 | mamAB | Related | ||
| mamI | amb0962 | mamAB | Specific | Vesicle, (Chain Formation?) | >berkeley 2010: Loss causes no membrane formation, is localized onto chains |
| mamE | amb0963 | mamAB; mam Islet | Related | >Membrane-bound serine protease required for magnetite formation; might control the localization of other magnetosome proteins | |
| mamJ | amb0964 | mamAB; mam Islet | Specific | Chain Formation | >Proper magnetosome chain organization/assembly |
| mamK | amb0965 | mamAB; mam Islet | Related | Chain Formation | >required for proper magnetosome chain organization; *bacterial actin-like cytoskeleton protein required for proper alignment of the magnetosomes in a chain, shown to localize the mamI |
| mamL | amb0966 | mamAB; mam Islet | Specific | Vesicle, biomineralization | >berkely 2010: Crucial to mangneosome membrane creation, shown to be spread across the cell membrane and sometimes forms lines |
| mamM | amb0967 | mamAB | Related | >biomineralization, involved in iron transport, magnetite nucleation, or establishement of the proper chemical enviornment for magnetite synthesis in the magnetosome | |
| mamN | amb0968 | mamAB | Related | >biomineralization, involved in iron transport, magnetite nucleation, or establishement of the proper chemical enviornment for magnetite synthesis in the magnetosome | |
| mamO | amb0969 | mamAB | Related | >biomineralization, involved in iron transport, magnetite nucleation, or establishement of the proper chemical enviornment for magnetite synthesis in the magnetosome | |
| mamP | amb0970 | mamAB | Related | Biomineralization | >berkeley 2010: loss causes weak magnetic response, with large but fewer crystals |
| mamA | amb0971 | mamAB | Related | >Required for magnetosome activation; activation of vessicles | |
| mamQ | amb0972 | mamAB; mam Islet | Related | >ORF; formation/maintenance of magnetosome membranes | |
| mamR | amb0973 | mamAB | Specific | Chain formation, Biomineralization | >ORF; plays a role in controlling both particle number and size of magnetite cyrstals |
| mamB | amb0974 | mamAB | Related | Vesicle, Biomineralization | >indirect role in magnetosome membrane invagination and biomineralization; magnetosome compartment formation |
| mamS | amb0975 | mamAB | Specific | ||
| mamT | amb0976 | mamAB | Specific | Biomineralization | >magnetite crystal growth; participates in different steps during magnetite synthesis |
| mamU | amb0977 | mamAB | Related | ||
| mamV | amb0978 | mamAB | N/A |
Using our two genes of interest, we created C-terminal sfGFP fusions so we could track the localization of each gene separately within E.coli.
| Strain AMB-1 | E.coli | |
|---|---|---|
| mamK-sfGFP | Cell 2 | Cell 3 |
| mamI-sfGFP | Cell B | Cell C |
The results we obtained with our sfGFP fusions inside E.coli were comparable to those done through other studies in the host organism Magnetospirillum magneticum. In both images
 "
"