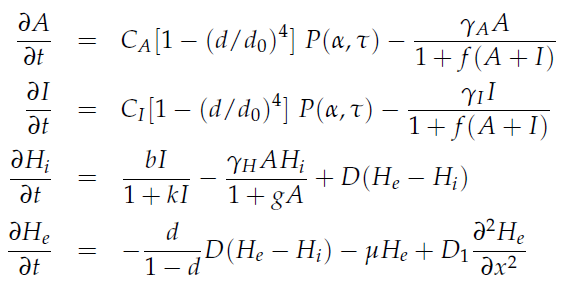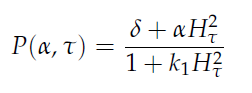Team:Wageningen UR/Project/ModelingProj1
From 2011.igem.org
(→Modeling synchronized oscillations) |
|||
| Line 31: | Line 31: | ||
| - | [[File:mainproject01.png]] | + | [[File:mainproject01.png|center]] |
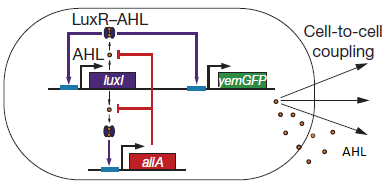
'''Fig.1:''' ''Basic oscillating genetic circuit as published by Danino & Hasty.'' | '''Fig.1:''' ''Basic oscillating genetic circuit as published by Danino & Hasty.'' | ||
Revision as of 20:50, 11 September 2011
 "
"