Team:Wageningen UR/Project/CompleteProject2Description
From 2011.igem.org
Complete Project Description
Introduction
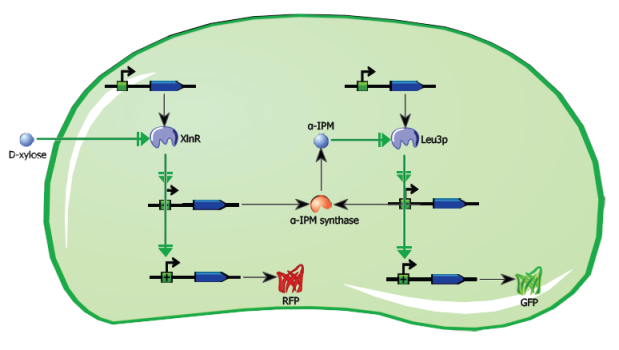
The aim of this project is to design and implement a sustained, self-propagating intercellular signalling system in the hyphal fungus Aspergillus nidulans. After initial induction, a genetic feed-forward loop will activate, resulting in the production of a signalling molecule. This molecule will diffuse through the septum into neighbouring cells, where it will enter the nuclei, and activate a transcription factor. This transcription factor will in turn activate the feed-forward loop, increasing the production of the signalling molecule and thereby allowing the signal to propagate throughout the hyphal network. This process will be visualized using fluorescent reporter proteins.
Mechanism
The initial activation of the circuit will be done by application of D-xylose to part of a hypha. D-xylose will enter the nucleus and bind to a transcription factor regulating the expression of red fluorescent protein and alpha-isopropylmalate-synthase via the XlnA:goxC promoter sequence. Thus the activation of this promoter region will cause RFP to be produced, as well as alpha-IPM-synthase, which will in turn synthesize alpha-IPM. Alpha-IPM will then diffuse through the septa and into the nuclei of neighbouring cells, where it will bind to the heterologous transcription factor Leu3p and activate the alpha-IPM feed-forward loop. Activated leu3p will also start the transcription of a GFP protein.
Because alpha-IPM is an intermediate in the organism’s Leucine biosynthesis pathway, it is necessary to use a Δa-IPMS A. nidulans strain to avoid spontaneous auto-induction of the system.
 "
"