Team:UNAM-Genomics Mexico/Notebook/nifH
From 2011.igem.org
(Created page with "{{:Team:UNAM-Genomics Mexico/Templates/Modeling| content= abi/hw }}") |
m (changed title) |
||
| (10 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:UNAM-Genomics Mexico/Templates/Modeling| content= | {{:Team:UNAM-Genomics Mexico/Templates/Modeling| content= | ||
| + | __NOTOC__ | ||
| + | |||
| + | |||
| + | |||
| + | =nifH and Anderson Promoters Logbook - ''Rhizobium etli'' kit= | ||
| + | |||
| + | ---- | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | The objective of this section is to explain the experimental strategies and activities that we undertook in order to achieve the following objectives: | ||
| + | |||
| + | *Characterization of the promoter region of the nifHa gene of ''Rhizobium etli''. The characterization involves describing the strength of this promoter region by comparing the expression GFP and RFP reporters under this promoter to the expression from a reference promoter from the iGEM Registry of parts, J23101. | ||
| + | |||
| + | |||
| + | *Characterization of the Anderson promoter collection for use in Rhizobia species. The aim is to describe the strength of the promoters that constitute this collection in our chasis, ''Rhizobium etli''. | ||
| + | |||
| + | |||
| + | The activities are organized by weeks (monday to sunday); we mention the general remarks for the activities that we performed. | ||
| + | |||
| + | |||
| + | __TOC__ | ||
| + | |||
| + | |||
| + | |||
| + | ==Week: June 13 - June 19== | ||
| + | |||
| + | This week we amplified the promoter region from the nifHa gene of ''Rhizobium etli''. We amplified this region by using primers that contained the prefix and suffix sequences in upper and lower primers respectively so that when we amplified the promoter region, we would end up with the prefix and suffix flanking the promoter region. We cleaned the PCR product and later assessed the size of the amplified product by gel electrophoresis. We saw that the size of amplified region did correspond to the size of the promoter region which is 252 bp (foto impresa). | ||
| + | |||
| + | |||
| + | ==Week: June 27 - July 1st== | ||
| + | |||
| + | This week we started by transforming four parts from the iGEM Spring 2011 Distribution, parts: J23101, J23102, E0240, and I20260. Parts J23101 and J23102 belong to the Anderson promoter collection, E0240 is a GFP generator, and I20260 is a measurement device for the promoter J23101. We achieved successful transformation for J23101, J23102, and E0240. We did these transformations to prove if the protocol we were going to use was fine. Since we had successful results, we continued with the transformation of the rest of the promoters in the Anderson promoter collection. | ||
| + | |||
| + | |||
| + | ==Week: July 4 - July 8== | ||
| + | |||
| + | We transformed competent cells with parts J23109, J23112, J23113, J23114, J23115 and I20260 because the first transformations performed with these parts were not successful. Unfortunately not all of these transformations were successful so we repeated them for a third time, this time increasing the amount of DNA with which we performed the transformation and at last we got good results. By the end of this week we has transformed competent bacteria with every part from the Anderson promoter collection as well as parts I20260 and E0240. We divided the Anderson promoter collection in two: the first part includes parts J23100 to J23111 and the second part includes parts J23112 to J23119. | ||
| + | |||
| + | |||
| + | ==Week: July 11- July 15== | ||
| + | |||
| + | Once we had ''E. coli'' transformed with all of the parts that we wanted, we started to extract plasmids from both the first and the second part of the promoter collection as well as from parts E0240 and I20260. We did this because our focus was to obtain as much clean plasmid from each part so we could later digest them with EcoRI and PstI and isolate the parts from an agarose gel. | ||
| + | |||
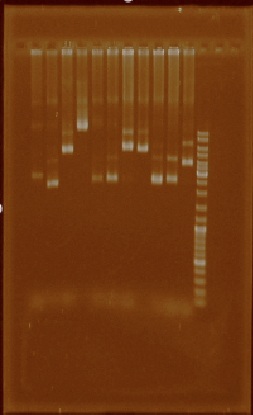
| + | [[File:Unamgenomicsfig1nifh.jpg|center|frame|Figure 1. Example of the low-quality plasmid extractions]] | ||
| + | |||
| + | |||
| + | Unfortunately most of the plasmid extractions didn’t result in clean plasmid (Figure 1). | ||
| + | We speculated this could have been a result of poor mini-prep solutions, so we made new ones. | ||
| + | |||
| + | |||
| + | ==Week : July 18- July 22== | ||
| + | |||
| + | We spent this week trying to obtain clean plasmids from the parts in the promoter collection, I20260, and E0240. Since we needed enough plasmid to later extract the band of interest from a low-melting point agarose gel we concentrated on getting the most and cleanest plasmids from the extractions, however this was resulting difficult as we still had trouble obtaining pure plasmid from the mini-preps. | ||
| + | |||
| + | |||
| + | ==Week: July 25- July 29== | ||
| + | |||
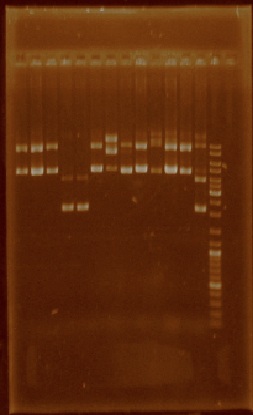
| + | This week we selected certain plasmid extractions which we thought were clean enough to proceed, these parts were: J23101 through J23110, J23113, J23114, and J23116 (Figure 2). The parts for which we still didn’t have clean or enough plasmid were E0240, I20260, J23100, J23105, J23106, J23111, J23112, J23115, J23117, J23118, and J23119, so we performed plasmid extraction for these parts once again. | ||
| + | |||
| + | [[File:Unamgenomicsfigure2nifh.jpg|center|frame|Figure 2. Plasmid extractions from J23101, J23102, J23103, J23104, J23105, J23106, J23107, J23108, J23109, J23110, J23113, J23114, and J23116 (the order of the lanes corresponds to the order of the above mentioned parts)]] | ||
| + | |||
| + | |||
| + | ==Week: August 1 - August 5== | ||
| + | |||
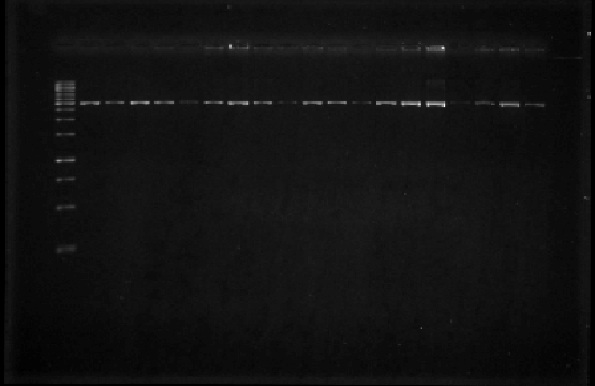
| + | In order to assess the sizes of the plasmids that we had selected last week, we digested them with EcoRI, then ran them on a 1% agarose gel (Figure 3). We then digested the plasmids with EcoRI and after this with PstI to be able to obtain the part of interest from an agarose gel. However, not all of the digestion reactions were successful so we spent this week further digesting the plasmids with EcoRI and PstI. | ||
| + | |||
| + | [[File:Unamgenomicsfig3nifh.jpg|center|frame|Figure 3. Digestion of the extracted plasmids with EcoRI. The bands correspond to a plasmid of ~3000 bp in size]] | ||
| + | |||
| + | |||
| + | ==Week: August 8 - August 12== | ||
| + | |||
| + | This week we finally had all of the plasmids pure enough to proceed with the digestions with EcoRI and PstI of the plasmids that hadn’t been extracted purely enough until this week. However not all of the digestions of the plasmids were resulting well. We decided to amplify the parts by PCR reactions instead of extracting the parts from a low-melting point agarose gel. | ||
| + | |||
| + | |||
| + | ==Week: August 15 - August 19== | ||
| + | |||
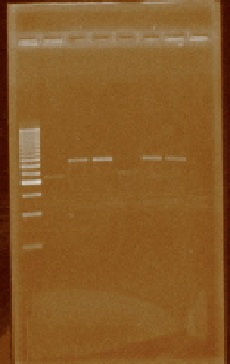
| + | We received the oligos to amplify regions within the iGEM prefix and suffix. We performed PCR reactions using the plasmids from parts E0240, J23100, J23105, J23106, J23111, J23112, J23115, J23117, J23118, and J23119 (Figure 4). We also amplified the I20260 part (foto 18 agosto). After amplifying the parts of interest we cleaned them by using a PCR product purification kit. | ||
| + | |||
| + | [[File:Unamgenomicsfig4nifh.jpg|center|frame|Figure 4. Gel with the PCR products from parts E0240, J23100, J23105, J23106, J23111, J23112, J23115, J23117, J23118. The lanes correspond to the PCR products in the order in which they were mentioned above. Part E0240 is ~925 bp in length due to the amplification of the prefix and suffix sequences; as for the promoter parts, their size is ~1000 bp in length also due to the prefix and suffix sequences.]] | ||
| + | |||
| + | |||
| + | ==Week: August 22 - August 26== | ||
| + | |||
| + | Since we had previously obtained pure plasmid for the part E0240, the GFP generator part with which we plan to de part of the promoter characterizations, and already had amplified the nifH promoter region, we decided to ligate the nifH promoter region to the E0240 plasmid by using the standard assembly method. To do this, we digested the nifH PCR product with EcoRI and SpeI and the E0240 plasmid with EcoRI and XbaI. Afterwards we dephosphorylated the 5’ ends in the digested E0240 and ligated it to the previously digested nifH promoter region. Transformation of ''E. coli'' competent cells was later performed with the products of the ligation reactions. | ||
| + | |||
| + | |||
| + | ==Week: August 29 - September 4== | ||
| + | |||
| + | We extracted plasmid from the colonies observed from the ligation of the nifH promoter region with the E0240 plasmid backbone (Figure 5). We digested the extracted plasmids from this ligation to check if the plasmid was the size we were expecting (four of the six plasmids were about the sized we had predicted). | ||
| + | |||
| + | Also as a part of our plans of characterizing the promoter region of nifH, we want to exchange the promoter from part J23101 for our promoter and thus drive the expression of a RFP reporter gene under the the nifH promoter. To achieve this, first we digested plasmid J23101 with EcoRI and SpeI in order to remove the original promoter from this plasmid. The plan is to introduce the nifH promoter to the linearized J23101 plasmid backbone by digesting the nifH promoter with EcoRI and SpeI as well. For this purpose we digested the nifH promoter with EcoRI and SpeI. We performed the ligation of the digested parts: nifH promoter region and J23101 plasmid backbone. | ||
| + | |||
| + | [[File:Unamgenomicsfig5nifh.jpg|center|frame|Figure 5. Linearized plasmids from the plasmid extractions of the ligation of the promoter region of nifHa and E0240 part as well as its plasmid.]] | ||
| + | |||
| + | This week we rekindled efforts on achieving the functional characterization of the promoters that constitute the Anderson promoter collection in our chasis ''Rhizobium etli''. We amplified the rest of the promoter parts that hadn’t been amplified before (parts J23101, J23102, J23103, J23104, J23107, J23108, J23109, J23110, J23113, J23114, and J23116). | ||
| + | |||
| + | |||
| + | ==Week: September 5 - September 11== | ||
| + | |||
| + | The PCR products from the promoter parts that we amplified last week, were digested with EcoRI and PstI in order to integrate them to our standardized plasmid. By running the digested promoter parts of the Anderson collection we were able to estimate the concentration of each promoter part. However we could no longer advance in this regard because we didn’t count with the standardized plasmid that we would use to transform ''E. coli'' and later conjugate with ''R. etli''. | ||
| + | |||
| + | The results of the transformations of ''E. coli'' with the ligations of the nifH promoter region and the J23101, promoter-less, plasmid backbone were odd. Since we had dephosphorylated the J23101 backbone plasmid we did not expect colonies in the control petri dish, which we saw. Moreover, there were colonies that had a distinctive light red color, which indicates that the RFP reporter in the J23101 plasmid is being actively transcribed; since the nifH promoter region is not recognized by the ''E. coli'' transcriptional machinery, there should be no expression of the RFP reporter. This could be explained if the digestion reaction to remove the promoter from the J23101 plasmid was not complete. However, among the colonies that grew in the ligation dishes, there were some that were transparent, so that led us to think that maybe those colonies had actually integrated the nifH promoter to the J23101 plasmid. | ||
| + | |||
| + | We decided to select 16 of these transparent colonies in order to see if they had the correct construction. It is from these colonies that we performed colony PCR. Of the 16 colonies, 14 actually amplified a DNA fragment of about the size of the ligation of the nifH promoter plus the J23101 reporter system (Figure 6). | ||
| + | |||
| + | [[File:Unamgenomicsfig6nifh.jpg|center|frame|Figure 6. PCR products amplified by a colony PCR method. This reaction was performed on colonies that were transparent because this is an indication that it doesn’t have the J23101 promoter. The amplified product bands correspond to the the size we expected of the ligatoin of the nifH prmoter and the RFP reporter system from the J23101 plasmid, which is approximately 1132 bp.]] | ||
| + | |||
| + | |||
| + | This week we also realized that the plasmids extracted from the cells transformed with the 'nifH promoter + J23101 plasmid' and 'nifH promoter + E0240 + E0240 plasmid' were not the right size (both plasmid extractions were about 2500 bp in size, when they ought to be 3194 and 3201 bp respectively) so we decided that we were going to repeat the ligations but this time, in order to avoid possible re-ligation problems, we were going to extract the parts we need directly from a low melting point agarose gel. This is why we extracted new plasmid from both the J23101 and E0240 parts. We also performed the digestion of the J23101 plasmid with EcoRI and SpeI. | ||
| + | |||
| + | |||
| + | ==Week: September 12 - September 18== | ||
| + | |||
| + | This week we performed the digestion of the plasmid of the E0240 part with EcoRI and XbaI in order to linearize this plasmid, and prepare it to be ligated with the nifH promoter. | ||
| + | |||
| + | As part of the characterization process of the nifH promoter region, we wanted to introduce the I20260 and J23101 parts into our standardized plasmid, so we cleaned the PCR products that had previously been obtained from these parts. | ||
| + | |||
| + | We also performed the extraction of J23101 reporter system (RBS + RFP + transcriptional terminators) and E0240 plasmids from a low-melting point agarose gel. After the completion of this task, we ran a gel to estimate the concentration of the E0240, “J23101” RFP reporter system, nifH promoter digested with EcoRI and SpeI parts, so we could ligate the necessary parts in order to obtain our reporting constructions. | ||
| + | |||
| + | |||
| + | ==Week: September 19 - September 25== | ||
| + | |||
| + | We performed the ligation reactions of the nifH promoter region with a) the E0240 part and its plasmid and b) J23101 reporter system and its plasmid. Once we plated ''E. coli'' cells transformed with these transformations, we only observed colonies in the plates with the ligation of the nifH promoter and the E0240 part. | ||
| + | |||
| + | It seemed that the PCR product from the I20260 part, after we cleaned it with a PCR product purification kit wasn’t right, so we amplified it once again, along with J23101 in order to have these parts ready to integrate into our standardized plasmid. | ||
| + | |||
| + | |||
| + | Back to the [[Team:UNAM-Genomics_Mexico/Project/RhizobialKit|Rhizobial kit Overview]] | ||
| - | |||
}} | }} | ||
Latest revision as of 21:37, 28 September 2011
nifH and Anderson Promoters Logbook - Rhizobium etli kit
The objective of this section is to explain the experimental strategies and activities that we undertook in order to achieve the following objectives:
- Characterization of the promoter region of the nifHa gene of Rhizobium etli. The characterization involves describing the strength of this promoter region by comparing the expression GFP and RFP reporters under this promoter to the expression from a reference promoter from the iGEM Registry of parts, J23101.
- Characterization of the Anderson promoter collection for use in Rhizobia species. The aim is to describe the strength of the promoters that constitute this collection in our chasis, Rhizobium etli.
The activities are organized by weeks (monday to sunday); we mention the general remarks for the activities that we performed.
Week: June 13 - June 19
This week we amplified the promoter region from the nifHa gene of Rhizobium etli. We amplified this region by using primers that contained the prefix and suffix sequences in upper and lower primers respectively so that when we amplified the promoter region, we would end up with the prefix and suffix flanking the promoter region. We cleaned the PCR product and later assessed the size of the amplified product by gel electrophoresis. We saw that the size of amplified region did correspond to the size of the promoter region which is 252 bp (foto impresa).
Week: June 27 - July 1st
This week we started by transforming four parts from the iGEM Spring 2011 Distribution, parts: J23101, J23102, E0240, and I20260. Parts J23101 and J23102 belong to the Anderson promoter collection, E0240 is a GFP generator, and I20260 is a measurement device for the promoter J23101. We achieved successful transformation for J23101, J23102, and E0240. We did these transformations to prove if the protocol we were going to use was fine. Since we had successful results, we continued with the transformation of the rest of the promoters in the Anderson promoter collection.
Week: July 4 - July 8
We transformed competent cells with parts J23109, J23112, J23113, J23114, J23115 and I20260 because the first transformations performed with these parts were not successful. Unfortunately not all of these transformations were successful so we repeated them for a third time, this time increasing the amount of DNA with which we performed the transformation and at last we got good results. By the end of this week we has transformed competent bacteria with every part from the Anderson promoter collection as well as parts I20260 and E0240. We divided the Anderson promoter collection in two: the first part includes parts J23100 to J23111 and the second part includes parts J23112 to J23119.
Week: July 11- July 15
Once we had E. coli transformed with all of the parts that we wanted, we started to extract plasmids from both the first and the second part of the promoter collection as well as from parts E0240 and I20260. We did this because our focus was to obtain as much clean plasmid from each part so we could later digest them with EcoRI and PstI and isolate the parts from an agarose gel.
Unfortunately most of the plasmid extractions didn’t result in clean plasmid (Figure 1).
We speculated this could have been a result of poor mini-prep solutions, so we made new ones.
Week : July 18- July 22
We spent this week trying to obtain clean plasmids from the parts in the promoter collection, I20260, and E0240. Since we needed enough plasmid to later extract the band of interest from a low-melting point agarose gel we concentrated on getting the most and cleanest plasmids from the extractions, however this was resulting difficult as we still had trouble obtaining pure plasmid from the mini-preps.
Week: July 25- July 29
This week we selected certain plasmid extractions which we thought were clean enough to proceed, these parts were: J23101 through J23110, J23113, J23114, and J23116 (Figure 2). The parts for which we still didn’t have clean or enough plasmid were E0240, I20260, J23100, J23105, J23106, J23111, J23112, J23115, J23117, J23118, and J23119, so we performed plasmid extraction for these parts once again.
Week: August 1 - August 5
In order to assess the sizes of the plasmids that we had selected last week, we digested them with EcoRI, then ran them on a 1% agarose gel (Figure 3). We then digested the plasmids with EcoRI and after this with PstI to be able to obtain the part of interest from an agarose gel. However, not all of the digestion reactions were successful so we spent this week further digesting the plasmids with EcoRI and PstI.
Week: August 8 - August 12
This week we finally had all of the plasmids pure enough to proceed with the digestions with EcoRI and PstI of the plasmids that hadn’t been extracted purely enough until this week. However not all of the digestions of the plasmids were resulting well. We decided to amplify the parts by PCR reactions instead of extracting the parts from a low-melting point agarose gel.
Week: August 15 - August 19
We received the oligos to amplify regions within the iGEM prefix and suffix. We performed PCR reactions using the plasmids from parts E0240, J23100, J23105, J23106, J23111, J23112, J23115, J23117, J23118, and J23119 (Figure 4). We also amplified the I20260 part (foto 18 agosto). After amplifying the parts of interest we cleaned them by using a PCR product purification kit.
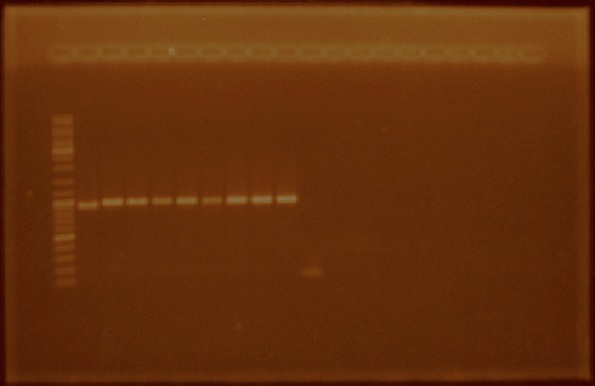
Week: August 22 - August 26
Since we had previously obtained pure plasmid for the part E0240, the GFP generator part with which we plan to de part of the promoter characterizations, and already had amplified the nifH promoter region, we decided to ligate the nifH promoter region to the E0240 plasmid by using the standard assembly method. To do this, we digested the nifH PCR product with EcoRI and SpeI and the E0240 plasmid with EcoRI and XbaI. Afterwards we dephosphorylated the 5’ ends in the digested E0240 and ligated it to the previously digested nifH promoter region. Transformation of E. coli competent cells was later performed with the products of the ligation reactions.
Week: August 29 - September 4
We extracted plasmid from the colonies observed from the ligation of the nifH promoter region with the E0240 plasmid backbone (Figure 5). We digested the extracted plasmids from this ligation to check if the plasmid was the size we were expecting (four of the six plasmids were about the sized we had predicted).
Also as a part of our plans of characterizing the promoter region of nifH, we want to exchange the promoter from part J23101 for our promoter and thus drive the expression of a RFP reporter gene under the the nifH promoter. To achieve this, first we digested plasmid J23101 with EcoRI and SpeI in order to remove the original promoter from this plasmid. The plan is to introduce the nifH promoter to the linearized J23101 plasmid backbone by digesting the nifH promoter with EcoRI and SpeI as well. For this purpose we digested the nifH promoter with EcoRI and SpeI. We performed the ligation of the digested parts: nifH promoter region and J23101 plasmid backbone.
This week we rekindled efforts on achieving the functional characterization of the promoters that constitute the Anderson promoter collection in our chasis Rhizobium etli. We amplified the rest of the promoter parts that hadn’t been amplified before (parts J23101, J23102, J23103, J23104, J23107, J23108, J23109, J23110, J23113, J23114, and J23116).
Week: September 5 - September 11
The PCR products from the promoter parts that we amplified last week, were digested with EcoRI and PstI in order to integrate them to our standardized plasmid. By running the digested promoter parts of the Anderson collection we were able to estimate the concentration of each promoter part. However we could no longer advance in this regard because we didn’t count with the standardized plasmid that we would use to transform E. coli and later conjugate with R. etli.
The results of the transformations of E. coli with the ligations of the nifH promoter region and the J23101, promoter-less, plasmid backbone were odd. Since we had dephosphorylated the J23101 backbone plasmid we did not expect colonies in the control petri dish, which we saw. Moreover, there were colonies that had a distinctive light red color, which indicates that the RFP reporter in the J23101 plasmid is being actively transcribed; since the nifH promoter region is not recognized by the E. coli transcriptional machinery, there should be no expression of the RFP reporter. This could be explained if the digestion reaction to remove the promoter from the J23101 plasmid was not complete. However, among the colonies that grew in the ligation dishes, there were some that were transparent, so that led us to think that maybe those colonies had actually integrated the nifH promoter to the J23101 plasmid.
We decided to select 16 of these transparent colonies in order to see if they had the correct construction. It is from these colonies that we performed colony PCR. Of the 16 colonies, 14 actually amplified a DNA fragment of about the size of the ligation of the nifH promoter plus the J23101 reporter system (Figure 6).
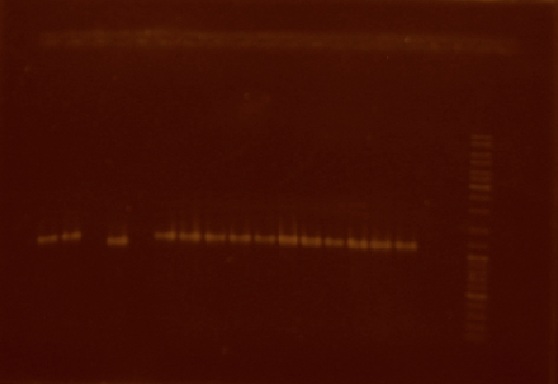
This week we also realized that the plasmids extracted from the cells transformed with the 'nifH promoter + J23101 plasmid' and 'nifH promoter + E0240 + E0240 plasmid' were not the right size (both plasmid extractions were about 2500 bp in size, when they ought to be 3194 and 3201 bp respectively) so we decided that we were going to repeat the ligations but this time, in order to avoid possible re-ligation problems, we were going to extract the parts we need directly from a low melting point agarose gel. This is why we extracted new plasmid from both the J23101 and E0240 parts. We also performed the digestion of the J23101 plasmid with EcoRI and SpeI.
Week: September 12 - September 18
This week we performed the digestion of the plasmid of the E0240 part with EcoRI and XbaI in order to linearize this plasmid, and prepare it to be ligated with the nifH promoter.
As part of the characterization process of the nifH promoter region, we wanted to introduce the I20260 and J23101 parts into our standardized plasmid, so we cleaned the PCR products that had previously been obtained from these parts.
We also performed the extraction of J23101 reporter system (RBS + RFP + transcriptional terminators) and E0240 plasmids from a low-melting point agarose gel. After the completion of this task, we ran a gel to estimate the concentration of the E0240, “J23101” RFP reporter system, nifH promoter digested with EcoRI and SpeI parts, so we could ligate the necessary parts in order to obtain our reporting constructions.
Week: September 19 - September 25
We performed the ligation reactions of the nifH promoter region with a) the E0240 part and its plasmid and b) J23101 reporter system and its plasmid. Once we plated E. coli cells transformed with these transformations, we only observed colonies in the plates with the ligation of the nifH promoter and the E0240 part.
It seemed that the PCR product from the I20260 part, after we cleaned it with a PCR product purification kit wasn’t right, so we amplified it once again, along with J23101 in order to have these parts ready to integrate into our standardized plasmid.
Back to the Rhizobial kit Overview
 "
"