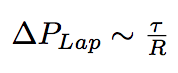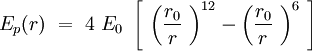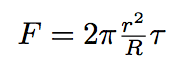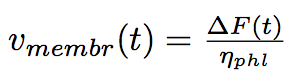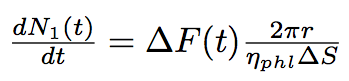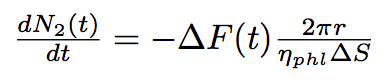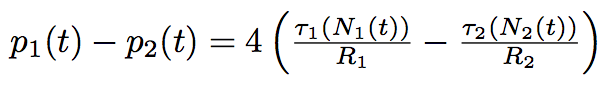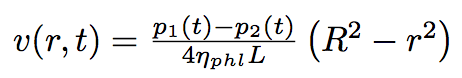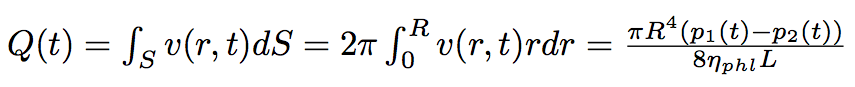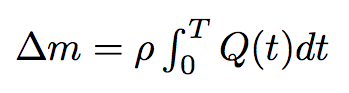Team:Paris Bettencourt/Modeling/Assisted diffusion
From 2011.igem.org

Assisted diffusion
Introduction to the model

|
Inspired by the experiments of Dubey and Ben-Yehuda we asked ourselves several questions: What kind of process could do this molecular transfer? How can we characterize it? It could be an active process, a passive diffusion or something else. Several arguments can be opposed to the active process hypothesis:
|
The question is :
We need to know if such a model can be designed starting from physical laws and if this model can explain quantitatively the transfer through the nanotubes. Due to its purely physical nature, our model can also shed some light as to the nanotube formation.
We managed to come up with an idea for such a process, and in this page we propose a new model that can possibly explain the speed of the molecule exchange through the nanotubes.
Two phenomena could lead to a pressure difference :
- The difference in membrane tension of two bacteria
- The osmotic pressure
Summary
The assisted diffusion model in 3 bullet points:
- Characteristic time of the process is about 100 ns
- The effect strongly depends on the initial phospholipid distribution on the membrane of two connected bacteria
- The phenomenon predicts the flux of only 0.1 % of the cytoplasme, not enough to explain the GFP experiment of the original paper
General physical concepts and hypotheses
How a connection between two bacteria might be established?
We do not know much about connections between bacteria. Let's suppose that the sugar envelope of one bacterium is somehow connected to another. Then inside this envelope, for several reasons, the inner pressure of bacteria can locally rise. This pressure can cause the formation of bulges growing from each bacteria inside the envelope. Their tubes could meet and even touch each other on the extremities.In this situation, the phospholipids on the ends of the tubes are rather unstable. Their hydrophobic parts are more exposed to water because of the high curvature on the extremities. So the energy-preferable state is to fuse.
In this case, there is no specific protein involved to do the fusion, so this explanation seems to be the compatible with reality.
In further paragraphs we will propose a version of the process happening after the membrane fusion.
Starting with a physical analogy
Imagine two bottles of gas connected by a tube, but closed by tape. The first one has a higher pressure than the second one. In the first one, there are a few molecules of another nature diluted in the gas. We follow these molecules.
When you open the tape ,the bottle with a higher pressure will come into equilibrium with the other one my moving a certain quantity of its particles through the tube in the direction of the second bottle. These moving molecules will drag with them the components diluted in the gas, and a few of these molecules will be transported to the other bottle.
From the analogy to biology
Of course, the cell is not a bag of liquid under pressure. The liquid pressure is equilibrated at both sides of the exterior membrane. The pressure we are dealing with, is not related to water or osmotic pressure (that is a "passive diffusion thermodynamical pressure"). There is a part of the cell we are not used to think about that undergoes a certain variabililty of pressure: the phospholipid membrane!
Let's evaluate the constraints that impact the membrane. As it is a Gram positive bacteria, the external sugar envelope imposes the shape of the bacteria. On the other hand, the osmotic pressure is pushing the membrane against the sugar wall. At every moment, the number of phospholipids inside the membrane is fixed, but this number can fluctuate depending on the phospholipid production. So we can pretend that our system is evolving through a series of quasi-equilibrium states.
 |
 |
When the cells "start the communication", a flow of phospholipids of the membrane can pass from the cell that has the highest membrane tension to the other one. To pass from one cell to another, phospholipids run around the tube.
The newly arrived phospholipids change the membrane tension of the bacterium, given our hypothesis of constant volume, which is justified by the sugar layer that "forbids" the bacterium to grow. As a consequence, they change the internal Laplace pressure of the bacterium.
where R is the radius of bacterium, τ is a membrane tension.
A simple analogy of clothesline can help to understand what is happening. You need to pull more your clothesline to put more clothes on it. When you pull the rope by two ends you create a tension. The bigger the tension, the more weight (pressure) you can put on the rope.
All of this will lead to establishing the pressure difference at the tube extremities and we will get a Poiseuille's flow. Constituents diluted in water will move from one cell to another unidirectionally and faster than by simple diffusion. This is a fast process that we have named the "assisted diffusion".
When membranes behave like 2D Van der Waals fluids
Trapped between the osmotic pressure and the cell wall, phospholipids are moving in a double layered environment. Their motion is constricted into a two dimensional motion, following the shape of the membrane. At the scale of the phospholipid, the motion can be approximated to a motion in 2D. Using statistical physics approach, we can say that the phase space has 4N dimensions: two dimensions of impulsion and two dimensions of coordinate for each particle (N is the number of particles in the system).
Can we describe the interaction between two phospholipids in the membrane? The question is complicated, but we will demonstrate that by describing properly the phospholipid, as it is reasonable to fit a Lennard-Jones potential energy to this interaction. The energy of binding is smaller and the speed of sliding of one phospholipid against another is slower than the internal vibration of the chemical bounds and the internal conformers rotation of the CH2 tails. Though, in Intermolecular Forces by Israelachvili, the author shows that a phospholipid trapped in a bilayer can be aproximated by a section of cone. The section of cones is an individual, and the others are sections of cone joining the previous one. The shape of the sections are giving the shape of the global structure.
 |
 |
Speaking in terms of energy, once we have these cones, it is reasonable to fit a Lennard-Jones potential to this interaction. If the cones are interpenetrating, a steric repulsion keeps the molecules apart, and the hydrophobic interaction that joins one lipid with the other acts as an attractive force keeping the coherence of the membrane. We will discuss later about the value of the two coefficients.
Is this phenomenon important? Back-of-the-envelope calculation
Let's start with a rough estimation of the flux transmitted through a nanotube. We assume that 5% of the surface area of one bacterium could be transmitted through the nanotube to a second bacterium. An order of magnitude of the transmitted surface is 10-14 m2. A nanotube of the radius 100 nm (that is the mean value of radius taken from the article) with this surface would be 0.5 µm long. The nanotubes observed in Dubey and Ben-Yehuda's experiments were about 100 nm to 1 µm.
Let's take for example a tube 0.2 µm long. That means that a total surface participating in the process is equivalent to 2.5*surfaces of the tube (the tube itself and the membrane parts adjacent to the tube). We can imagine the process to be split in two stages: the first is the establishment of contact between two bacteria, for that the equivalent of 1 surface of the tube will be used. The remaining 1.5 surface equivalents of the tube will be used to transport the liquid inside the tube, as it will be pulled along with the membrane. As the diameter of the tube is of order of 100 nm, we can neglect the complexity of the fluid movement inside. In our model we will consider the fluid to be moving with the membrane. So the volume of transmitted liquid is about 10-20 m3, whereas the cell volume is 5*10-18 m3.
We consider that some molecules are produced in the first cell and they are uniformly distributed. With 10'000 molecules in the first cell, we can expect about 200 molecules transmitted through one nanotube to another cell. That number of molecules seems to be sufficient for detection.
The membrane tension calculation
Considering one membrane a sphere of the radius R, on which the phospholipid double layer is uniformally distributed, we can find approximately a part of the whole sphere surface per one phospholipid :









Back to the classical physics
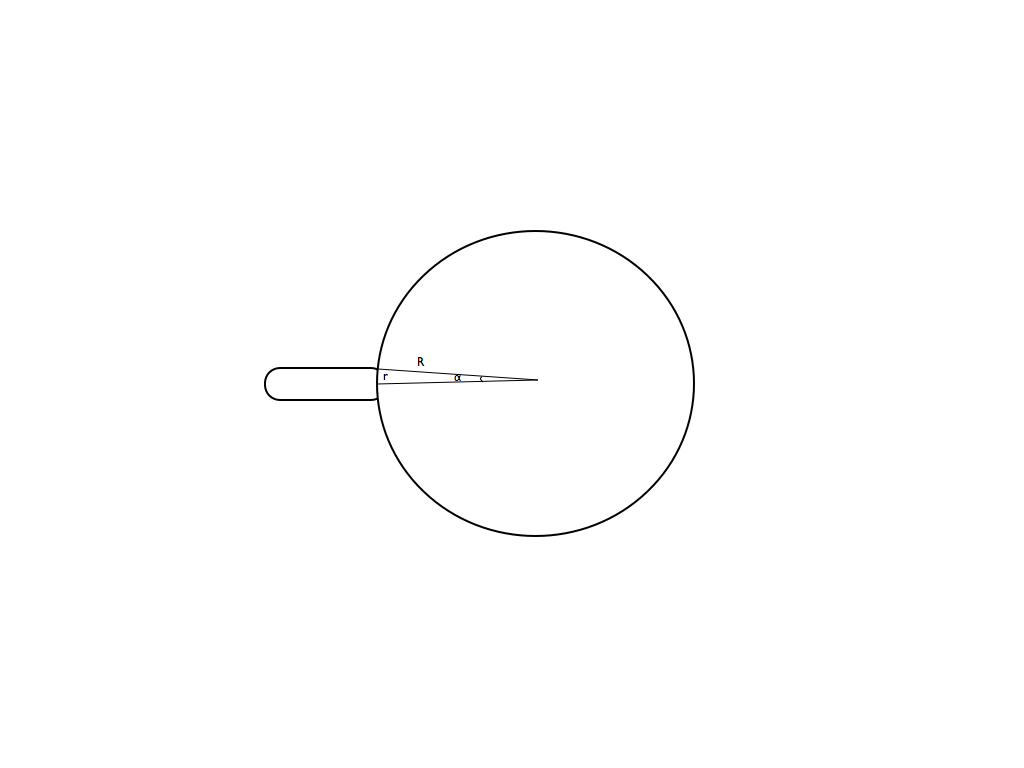
To deduce the force exercising on the tube from membranian pressure, please refer to an explanation figure below :
Here T is a surface tension, N is a resulting force of the area of the tube attachment to the bacterium, Δα or α is the half of the angle with a vertex in the center of the bacterium and between the tube extremities. Only this zone is giving a non-compensated contribution to the resulting force.
So the force exercising on the tube is given by the formula :
where r is the tube radius, R is the bacterium radius.
At first approximation we can consider the process a serie of quasi equilibrium states, that it is rather slow that acceleration of the membrane is zero. We find the speed of the tube membrane at every moment of time out of the second Newton's law :
where ΔF is the difference of the forces applied to the extremities of the tube.
The number of phospholipids in two bacteria will change according to these equations :
where N1, N2 are numbers of phospholipids on the membrane of the first and the second bacterium corrispondingly, ΔS is the surface per one phospholipid one the tube.
Cutting the whole period of time of our process into laps of time dt we should solve the Poiseuille equation each time reevoluating the difference of pressure :
The Poiseuille equation :
where L is the total length of the nanotube, r is the distance from the center line of the nanotube, R is the radius of the nanotube.
The volume flow of the liquid passed through the nanotube is :
By integrating the last formula in time we will get the mass of the liquid passed from one bacterium to another :
where T is the time of the process.
Getting good parameters
We have done several simulations where the system consisted of two spherical bacteria of radius R1 and R2, the nanotube of the length L and the radius r between them, and the number of phospholipids N1 and N2 on them.
The nanotube dimentions we have taken from the article published by Dubey and Ben-Yehuda [1] (r = 100 nm, L = 1 µm). The ideas about what values to take for R1, R2 and N1, N2 we got from the bionumber's site [2]. We have checked several sets of parameters :
- N1 = 2*106, N2 = 1*106, R1 = R2 = 0.5 µm;
- N1 = 1*107, N2 = 5*106, R1 = 2 µm, R2 = 1 µm;
The value of dynamic viscosity of phospholipid bilayer taken is 10 Pa.s, a bit more than what can be found for castor oil.
Analysing the results and conclusion
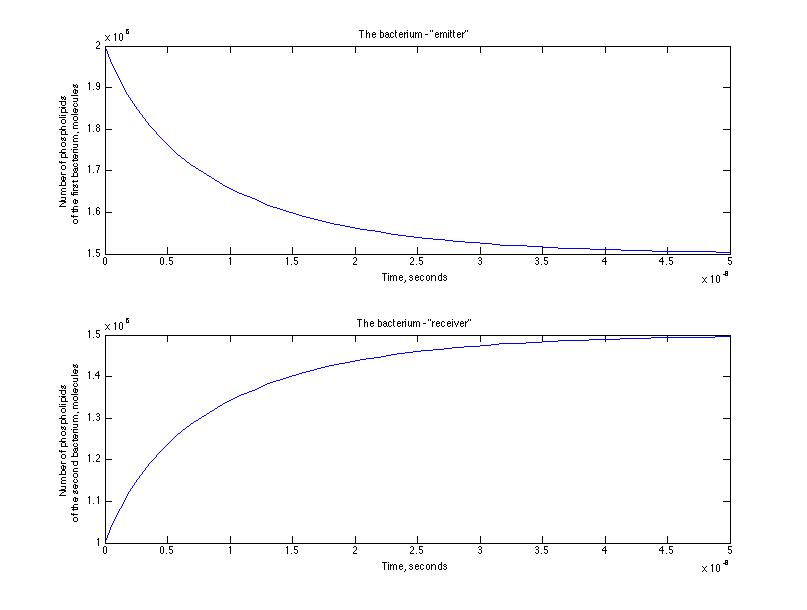
Here is the representation of the cinetic between the emitter and receiver - the number of phospholipids passed from the emitter to the receiver through the nanotube.
After a set of simulations we have estimated the characteristic time of the process and it was of the order of 10-8 seconds for the first simulation and 10 -6 seconds for the second one. That means that we were right at the beginning that an assisted diffusion process is much faster that a passive diffusion.
The masses transported are of the order of 10 -18 kg, which is an interesting result. In terms of the volume exchange : we can observe that about 10 -21 m 3 of liquid is transfert from the emitter to the receiver. 0,1 % of the emitter volume is transported due to an assisted diffusion. This doesn't seem much and it certainly doesn't entirely explain the original gfp diffusion experiment. It is nevertheless interesting to see such orders of magnitude.
Anyway this process can assist the passive diffusion in a way that if we have a lot of molecules in one bacterium that are localized near the membrane and nanotubes, it may accelerate the diffusion.
We still have to work on this model in order to tinker with some of the problem parameters. This might permit us to see a faster and more significant molecule transport through the nanotubes.
References
 "
"