Team:Paris Bettencourt/Experiments/tRNA diffusion
From 2011.igem.org
Danyel.lee90 (Talk | contribs) |
|||
| (29 intermediate revisions not shown) | |||
| Line 21: | Line 21: | ||
<h2>Parts and BioBricks construction</h2> | <h2>Parts and BioBricks construction</h2> | ||
| - | <b> | + | <b>Here is the cloning plan we followed for building our construction:</b> |
| + | |||
| + | <center> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <a href="https://static.igem.org/mediawiki/2011/8/87/1028_Cloning_plans_T7_em.png"><img src="https://static.igem.org/mediawiki/2011/8/87/1028_Cloning_plans_T7_em.png" width=550></a> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <a href="https://static.igem.org/mediawiki/2011/9/9b/1028_Cloning_plans_tRNA_rec.png"><img src="https://static.igem.org/mediawiki/2011/9/9b/1028_Cloning_plans_tRNA_rec.png" width=550></a> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | </center> | ||
| + | |||
<h2>Characterization of the tRNA amber</h2> | <h2>Characterization of the tRNA amber</h2> | ||
| Line 44: | Line 56: | ||
| - | <h3> | + | <h3>Fluorescence lamp</h3> |
<p>The cultures of strains having transformed both tRNA amber suppressor and GFP amber were strongly fluorescent, while the culture of the GFP amber alone as well as the culture of the colony without IPTG induction showed basal fluorescence. | <p>The cultures of strains having transformed both tRNA amber suppressor and GFP amber were strongly fluorescent, while the culture of the GFP amber alone as well as the culture of the colony without IPTG induction showed basal fluorescence. | ||
| Line 52: | Line 64: | ||
<html> | <html> | ||
| - | <h3> | + | <h3>Microscopy</h3> |
<h4>GFP amber</h4> | <h4>GFP amber</h4> | ||
| Line 100: | Line 112: | ||
|[[File:Trna iptg gfp amber fluo 4.jpg|350px|thumb|center|E-Coli at 37°C (gfp image)]] | |[[File:Trna iptg gfp amber fluo 4.jpg|350px|thumb|center|E-Coli at 37°C (gfp image)]] | ||
|} | |} | ||
| + | <p>The cultures of strains having transformed both tRNA amber suppressor and GFP amber were strongly fluorescent, while the culture of the GFP amber alone as well as the culture of the colony without IPTG induction showed basal fluorescence. | ||
| + | </p> | ||
| + | <html> | ||
| + | <h3>Spectrophotometry</h3> | ||
| + | </html> | ||
| + | <p>The same strains mentioned above were cultured overnight then introduced into a 96-well spectrophotometry plate. They were diluted directly in the wells to an OD of approximately 0.2. The green fluorescence and the OD600 were followed for 5hrs at 37°C. | ||
| + | </p> | ||
| + | [[File:GFPamberwithtRNATECANgraph1.jpg|700px|thumb|center]] | ||
| + | <html> | ||
| + | <h2>Characterization of the T7 amber in E.coli</h2> | ||
| + | <h3>Spectrophotometry</h3> | ||
| + | <p>The pT7_GFP control strain holds the following construct: pT7_RBS_GFP_tt.</p> | ||
| + | <p>The pT7_GFP:Pveg_T7amber strain holds, in addition to the previous construct, a T7 polymerase with two amber mutations under the control of a strong constitutive promoter.</p> | ||
| + | <br>The pT7_GFP:Pveg_T7amber:Phyperspank_tRNAamber is a E.coli strain holding the two previous constructs as well as a tRNA amber suppressor under the control of an IPTG inducible promoter: PHyperspank. The transformed plasmid also holds a <b><a href="https://2011.igem.org/Team:Paris_Bettencourt/Experiments/pHyperSpank">LacI gene</a></b> under the control of Pveg.</br> | ||
| + | |||
| + | <p>All strains were cultured overnight, then introduced into a 96-well spectrophotometry plate. They were diluted directly in the wells to an initial OD600 of 0.2. The green fluorescence as well as the OD600 were then followed for 3hrs at 37°C. | ||
| + | </p> | ||
| + | </html> | ||
| + | [[File:T7ambertRNAgraph1.jpg|700px|thumb|center]] | ||
| + | <p>The basal expression of GFP is higher in pT7_GFP:Pveg_T7amber than the pT7_GFP control due to the leaky expression of the T7 polymerase. </p> | ||
| + | <p>In the presence of the appropriate tRNA amber suppressor, the GFP expression increases significantly, reflecting the increase in the translation of functional T7 polymerase. | ||
| + | Therefore, the tRNA amber suppressor as well as the T7amber are fully functional with minimal leakage levels.</p> | ||
<html> | <html> | ||
| + | <p>The T7amber was also introduced into the same strain with the <b><a href="hhttps://2011.igem.org/Team:Paris_Bettencourt/Experiments/T7_diffusion">T7 autoloop</a></b>. </p> | ||
| + | </html> | ||
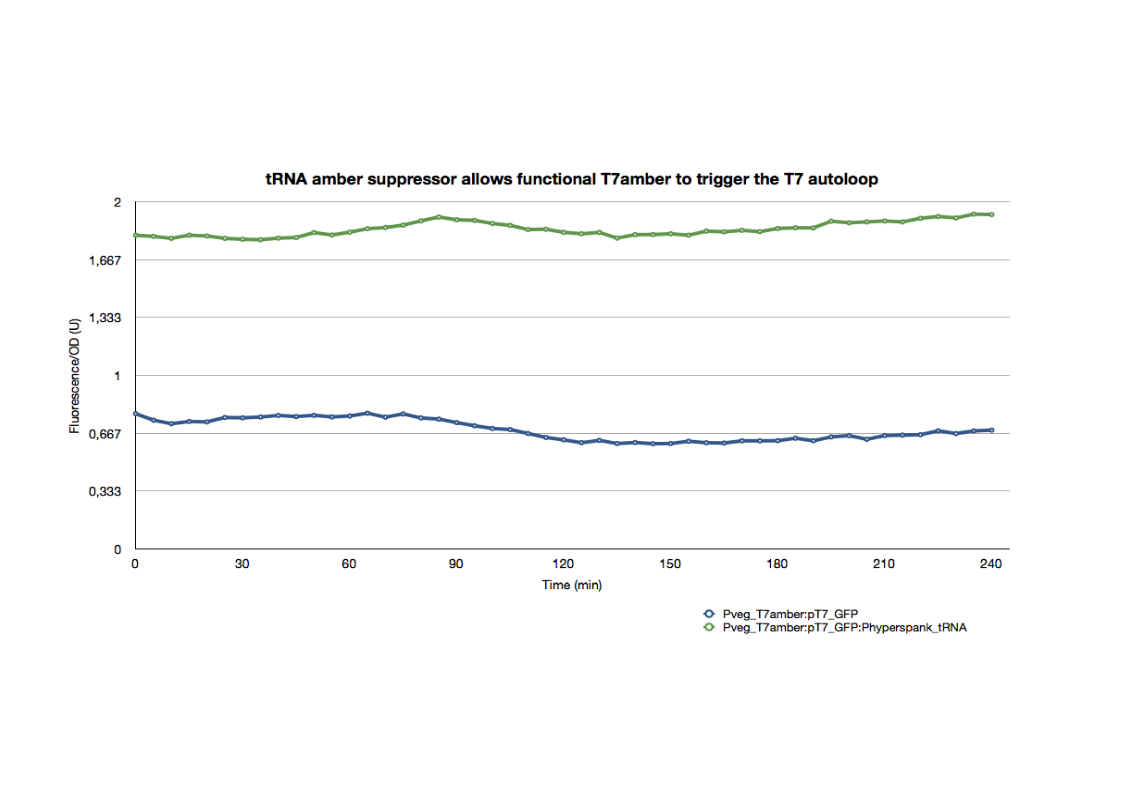
| + | <p>The GFP expression of this strain after an overnight culture was compared with that of a strain holding the same constructs in addition to the tRNA amber suppressor. The strains were introduced into a 96-well plate, diluted to an initial OD600 of approximately 0.1 directly in the wells. The green fluorescence and the OD600 were followed for 4hrs at 37°C.</p> | ||
| + | [[File:T7ambertRNAgraph2.jpg|700px|thumb|center]] | ||
| + | |||
| + | <p>The difference in the GFP expression is proof of a functional T7amber triggering the T7 autoloop when the appropriate tRNA amber suppressor is expressed. All final constructs for the tRNA system are functional in E.coli with behavioral profiles consistant with our expectations.</p> | ||
| + | |||
| + | <html> | ||
<!-- PAGE FOOTER -- ITEMS FROM COLUMN ! HAVE BEEN MOVED HERE -- RDR --> | <!-- PAGE FOOTER -- ITEMS FROM COLUMN ! HAVE BEEN MOVED HERE -- RDR --> | ||
Latest revision as of 03:37, 29 October 2011

Building and characterizing the tRNA amber design
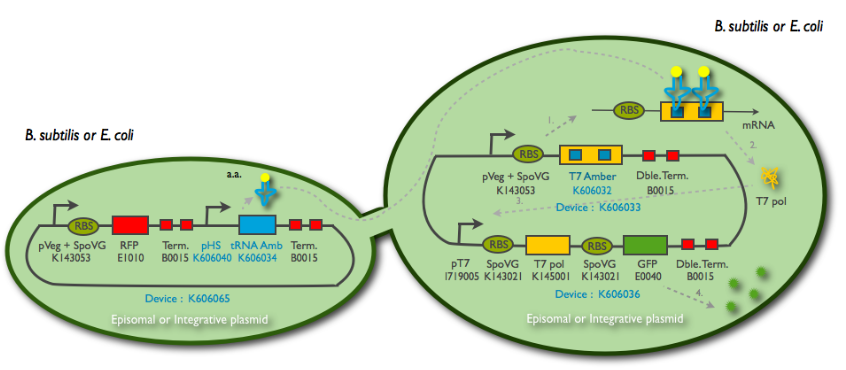
The tRNA amber diffusion design uses the concept of "amber" BioBricks, that is BioBricks that can not be properly translated without a tRNA associated with the amber stop codon and carrying a tyrosine amino-acid. We use an auto-amplification device base on this polymerase that we call T7 autoloop. This autoloop is also used in the T7 RNA polymerase diffusion design. We successfully BioBricked and characterized all our constructs for this design and made them available for the synthetic biology community. Our results are detailed below.
Abstract
Results for the tRNA amber diffusion design:
- We successfully BioBricked the tRNA amber emitter (BBa_K606041), the T7 amber receptor (BBa_K606033) and the T7 autoloop (BBa_K606036) constructs and sent them to the registry
- We characterized the tRNA amber in E.coli
- In order to characterize the tRNA amber, we successfully BioBricked and characterized a GFP amber gene (BBa_K606043)
- We saw that our constructs behave accordingly to the model we made
Design overview
Principles of the tRNA amber diffusion system
Parts and BioBricks construction
Here is the cloning plan we followed for building our construction:

Characterization of the tRNA amber
Protocol
In order to characterize the tRNA amber suppressor, as well as the pHyperspank, a special part was made: Pveg-SpoVG-GFP amber-TT.
A double transformation was done with minipreps of two plasmids of different antibiotic resistances : pSB1C3 holding the tRNA under the control of pHyperspank and pSB1AK3 holding the GFP with the amber codon under the control of a constitutive promotor. Two groups of cultures of each picked colony were made. One group was cultured in rich medium without IPTG whilst the other group was cultured in rich medium with IPTG (4mM in 10mL). A culture of the strain holding GFP amber alone was grown in parallel. The two tubes of each colony were compared along with the negative control (GFP amber alone). Cultures were then diluted for microscopy, glycerols, and future experiments. We used the following strains to characterize this system:
GFP amber Strain holding GFPmut3B, mutated with a single amber mutation under the control of the constitutive promotor Pveg, in the plasmid pSB1AK3 (AmpR).
tRNA amber suppressor Strain holding the tRNA amber suppressor, under the control of IPTG inducible strong promotor pHyperspank, in the plasmid pSB1C3 (CmR).
Double transformant Double transformant of the two previous constructs in their plasmids respectively.
Two groups of cultures are grown: with or without IPTG.
Direct observation under fluorescence lamp and microscopy was used to characterize this system.
We have also characterised the pHyperSpank promoter, compatible with B.subtilis and which is used in this system.
Fluorescence lamp
The cultures of strains having transformed both tRNA amber suppressor and GFP amber were strongly fluorescent, while the culture of the GFP amber alone as well as the culture of the colony without IPTG induction showed basal fluorescence.
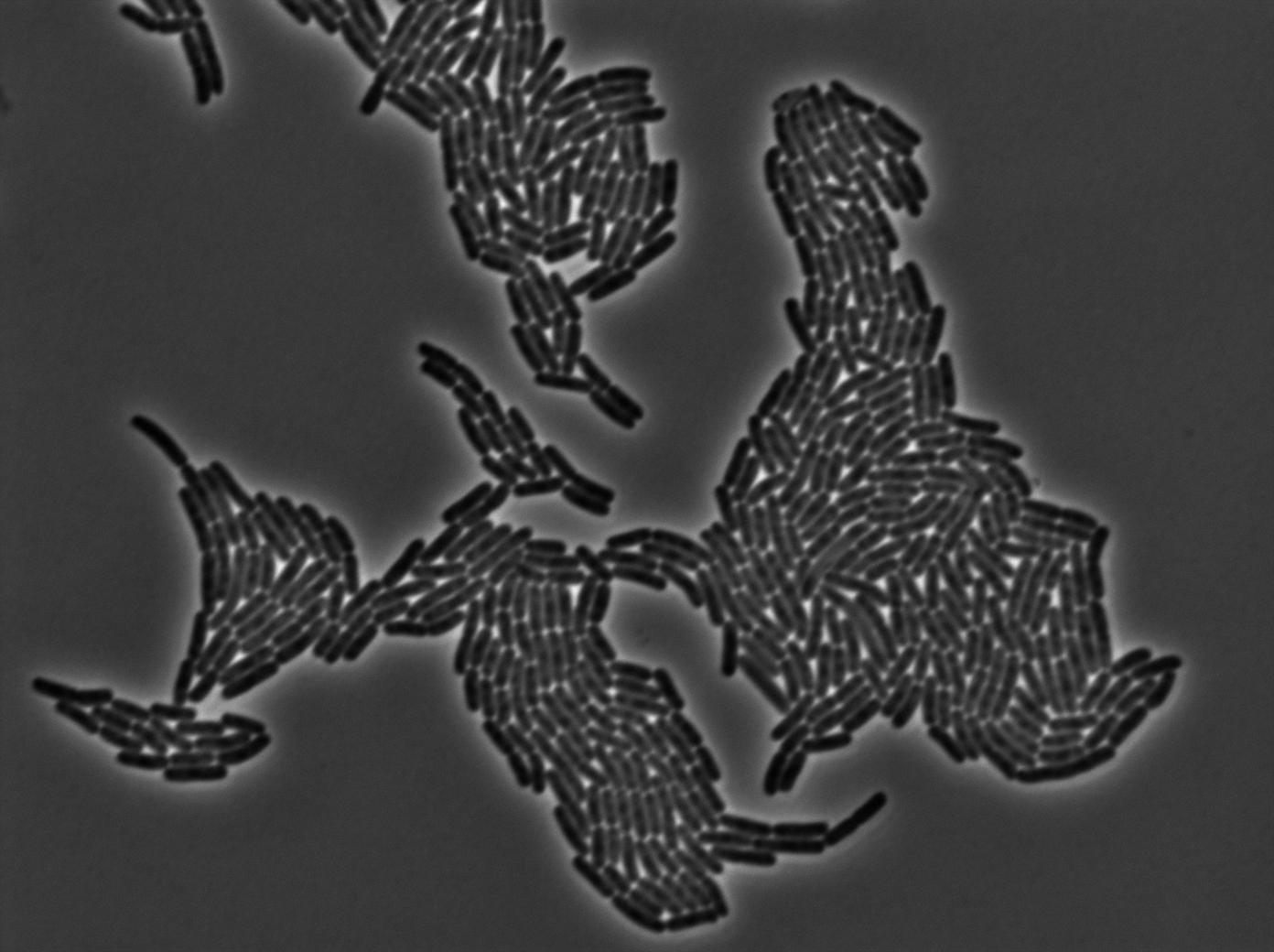
Microscopy
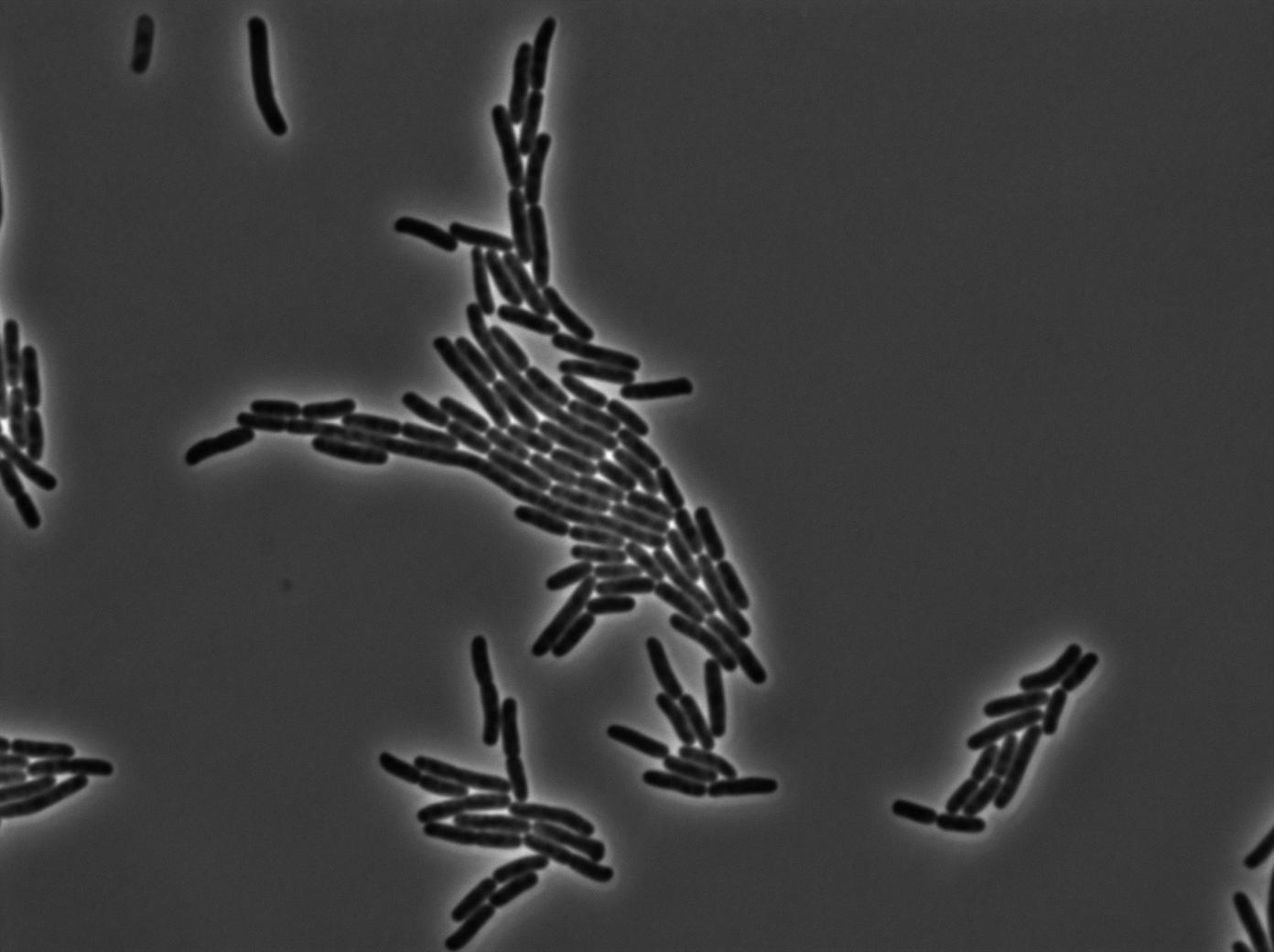
GFP amber
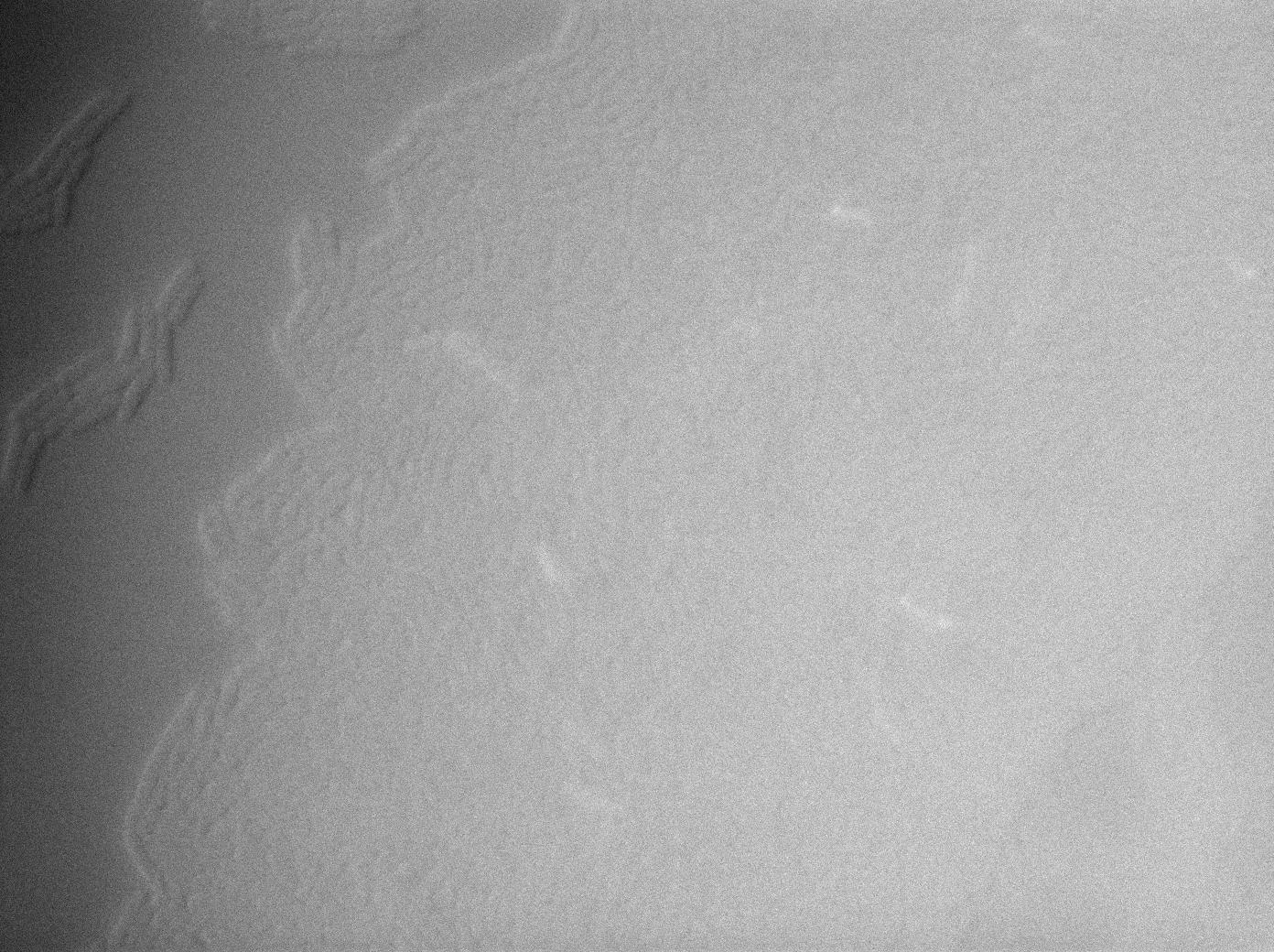
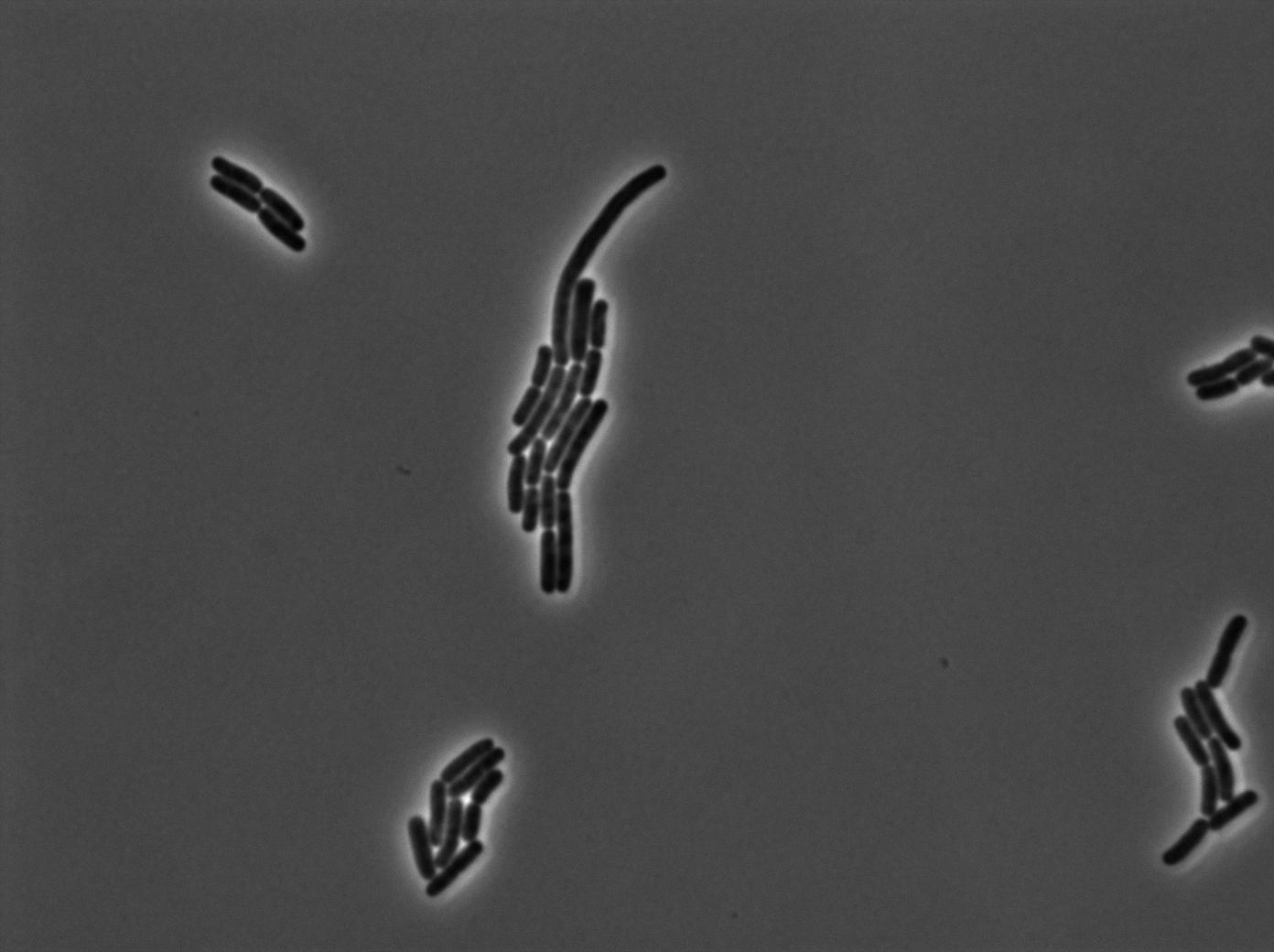
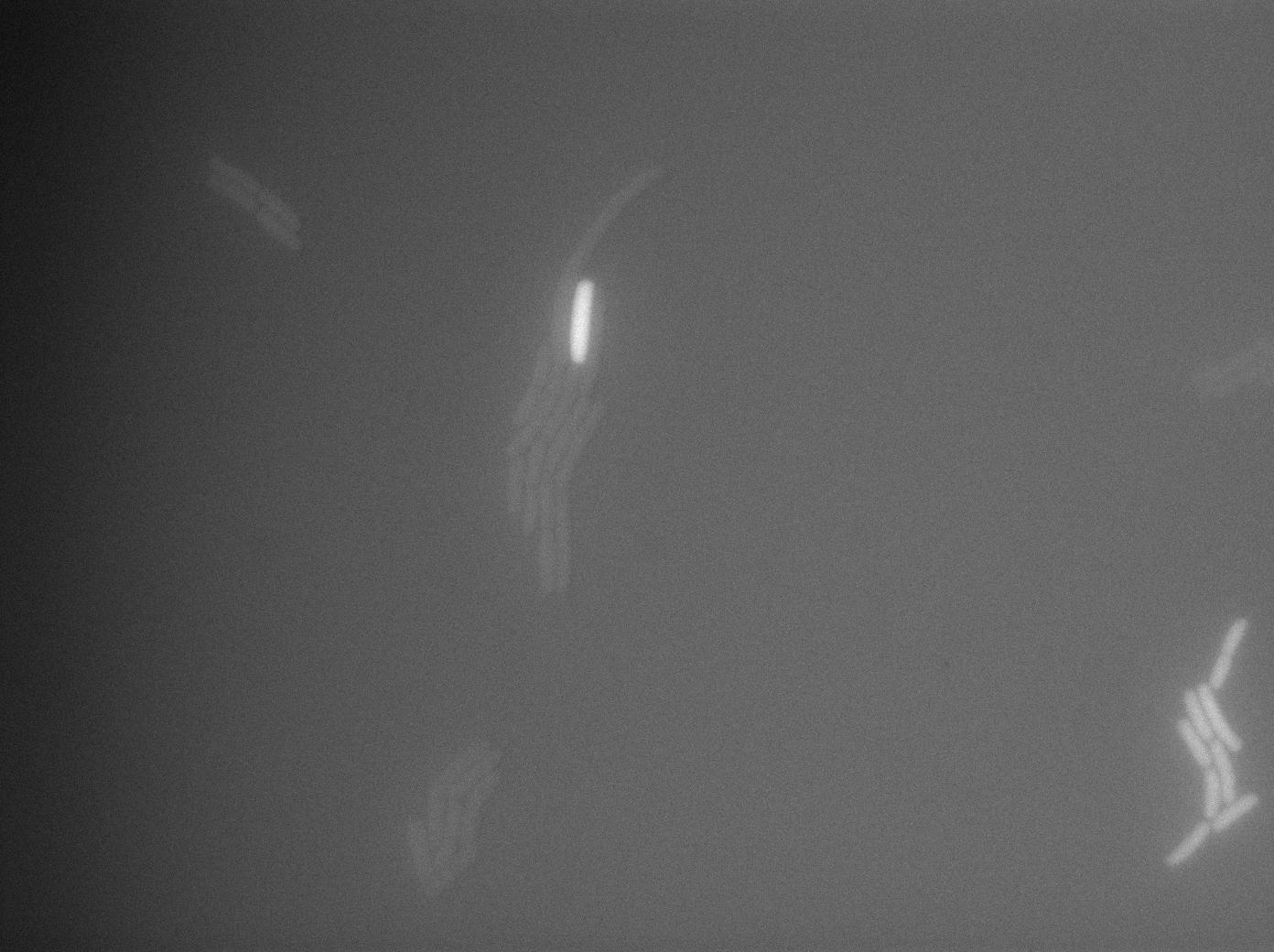
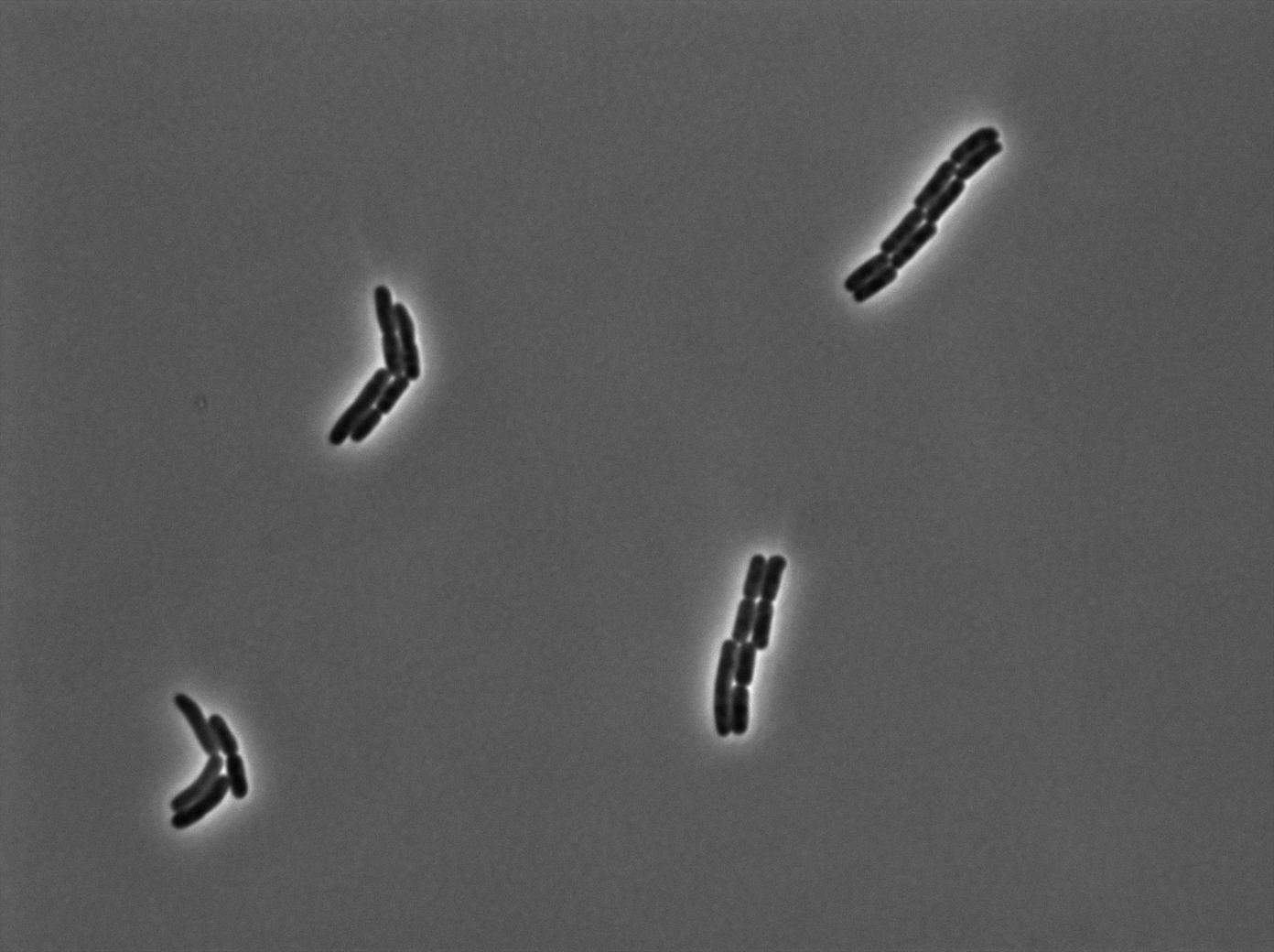
One of our strains contains only the plasmid with the GFP amber under the control of the constitutive promotor Pveg, without the amber suppressor tRNA. In this situation cells do not glow. This obervation constitutes a reliable negative control.
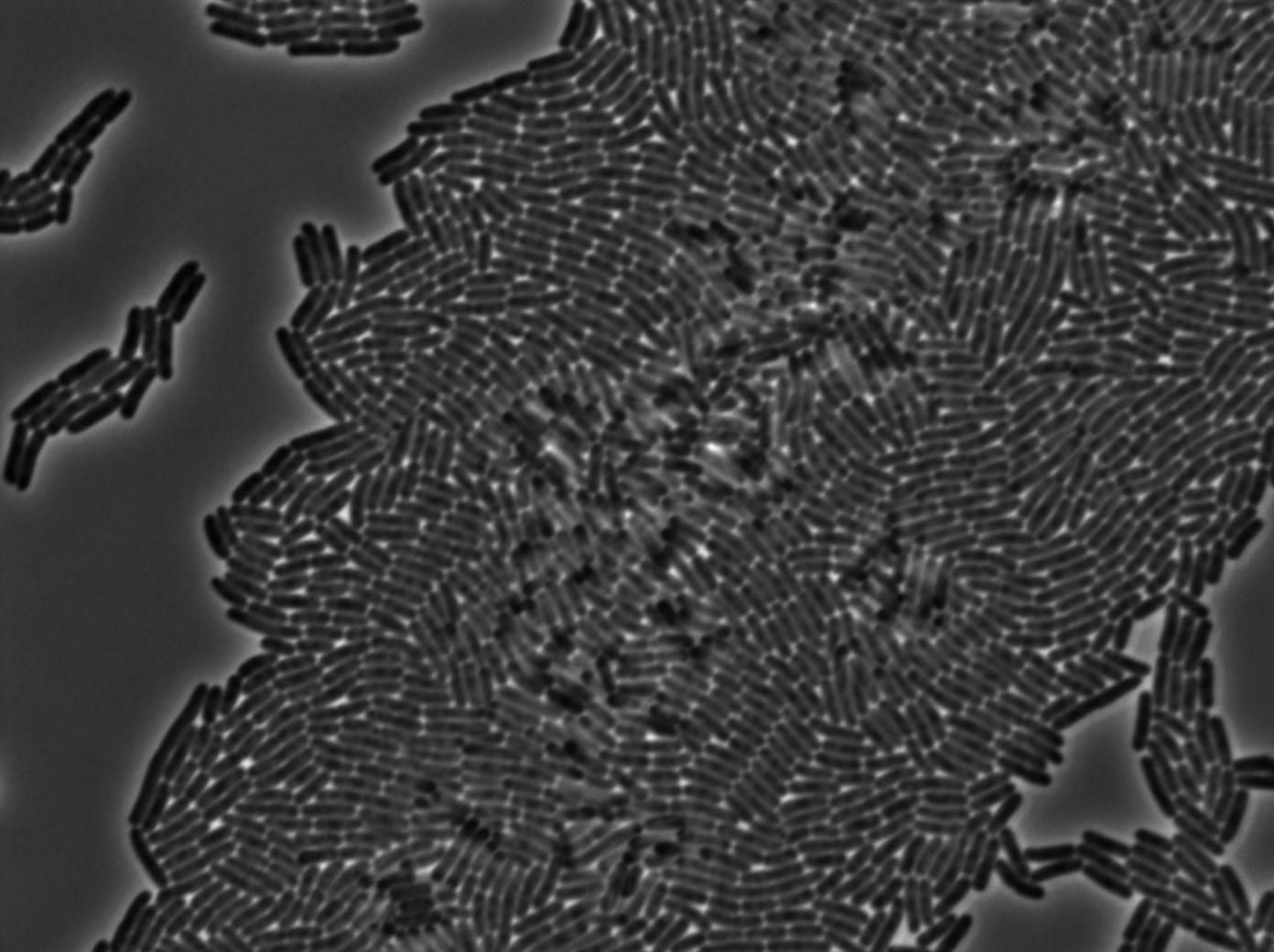
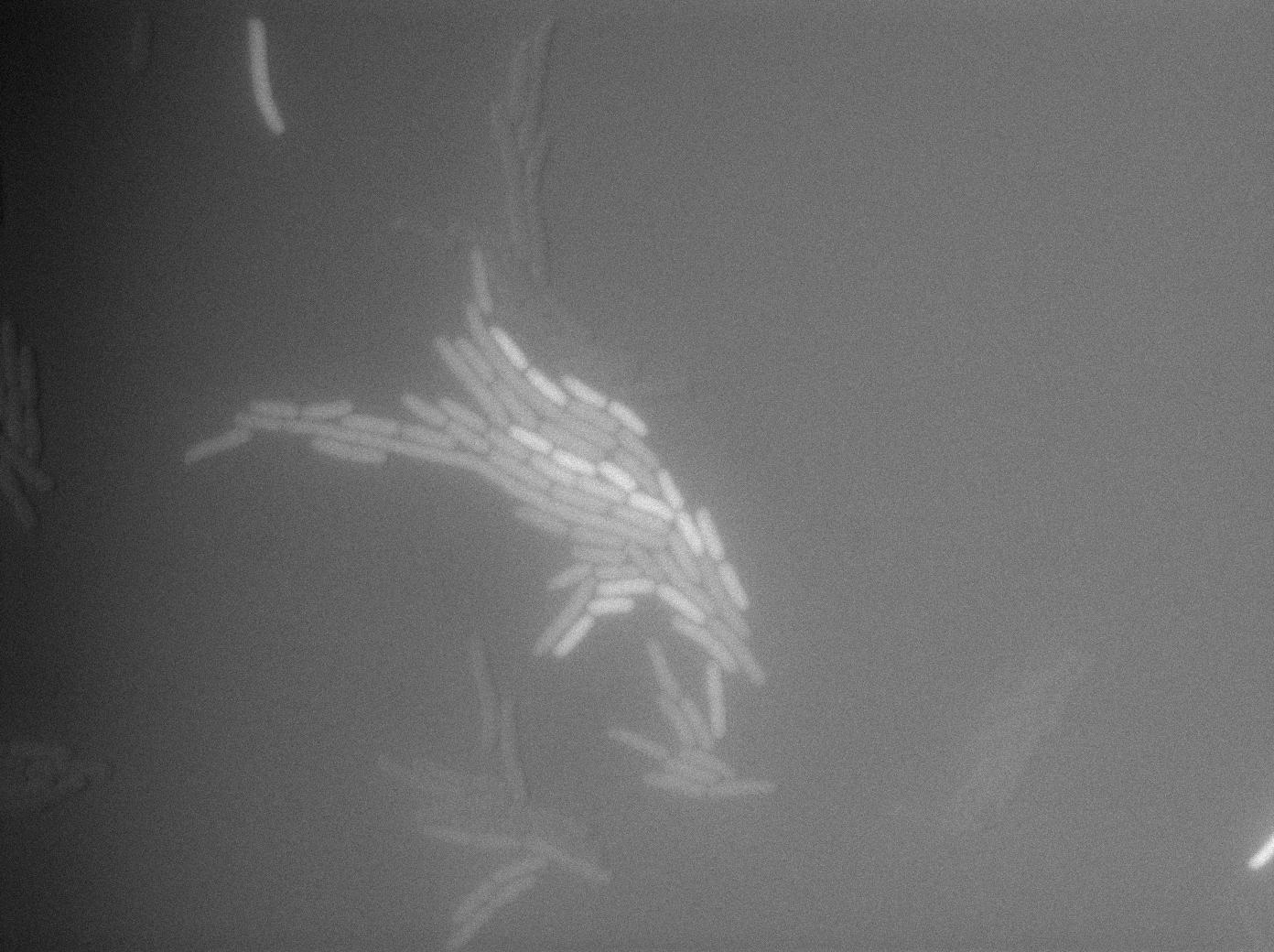
GFP amber with tRNA amber suppressor
The second strain contains the full construct, and the production of tRNA is tuned by pHyperspank which is activated by IPTG. In order to make a negative control, we plated them without inducing with IPTG. In this situation cells glow with a lower intensity than with the previous construct. We can also notice that some of the cells strongly leak.
GFP amber with tRNA amber induced by IPTG
The last result is given by testing the full construct with IPTG induced cells. Each cell glows with a very weak variability and the luminance is significant.
The cultures of strains having transformed both tRNA amber suppressor and GFP amber were strongly fluorescent, while the culture of the GFP amber alone as well as the culture of the colony without IPTG induction showed basal fluorescence.
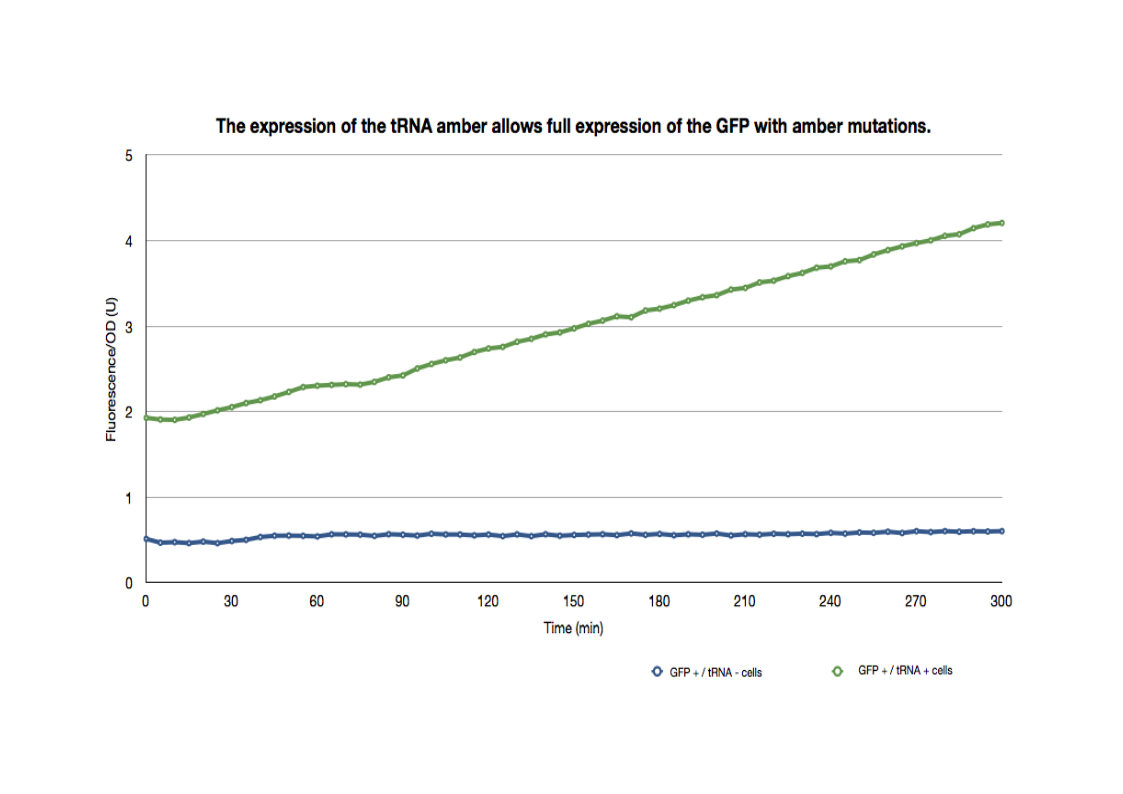
Spectrophotometry
The same strains mentioned above were cultured overnight then introduced into a 96-well spectrophotometry plate. They were diluted directly in the wells to an OD of approximately 0.2. The green fluorescence and the OD600 were followed for 5hrs at 37°C.
Characterization of the T7 amber in E.coli
Spectrophotometry
The pT7_GFP control strain holds the following construct: pT7_RBS_GFP_tt.
The pT7_GFP:Pveg_T7amber strain holds, in addition to the previous construct, a T7 polymerase with two amber mutations under the control of a strong constitutive promoter.
The pT7_GFP:Pveg_T7amber:Phyperspank_tRNAamber is a E.coli strain holding the two previous constructs as well as a tRNA amber suppressor under the control of an IPTG inducible promoter: PHyperspank. The transformed plasmid also holds a LacI gene under the control of Pveg.
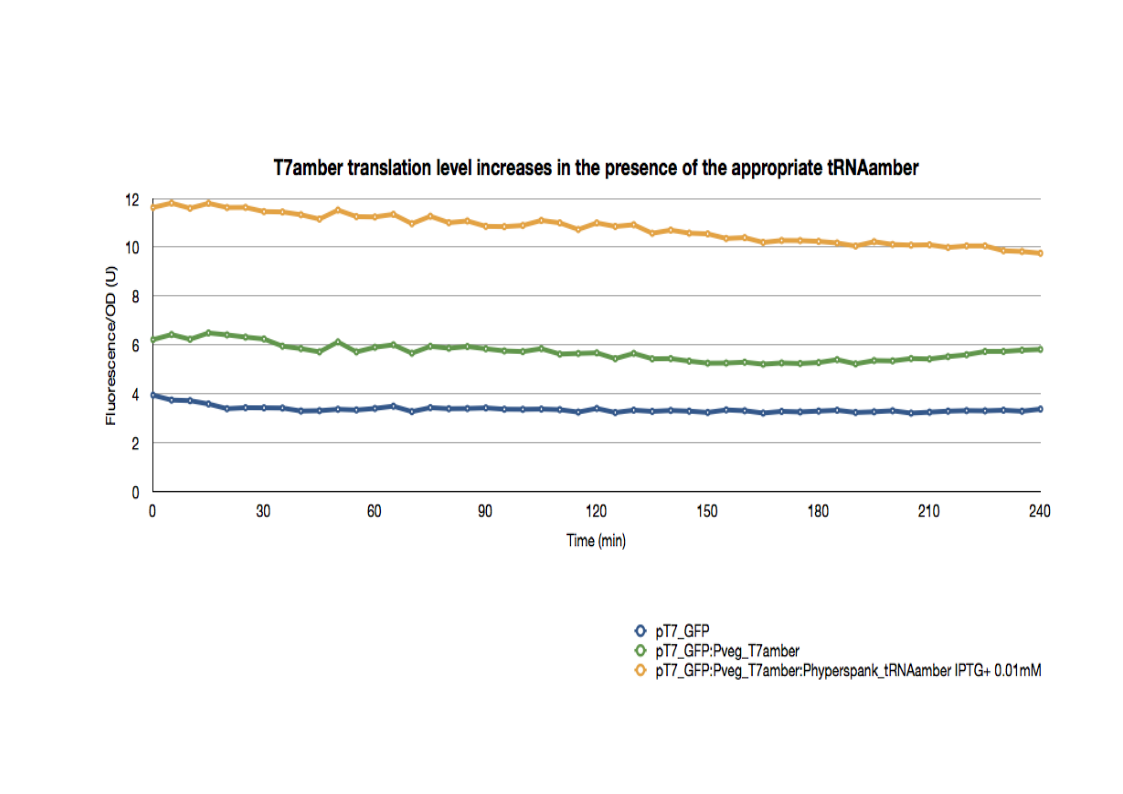
All strains were cultured overnight, then introduced into a 96-well spectrophotometry plate. They were diluted directly in the wells to an initial OD600 of 0.2. The green fluorescence as well as the OD600 were then followed for 3hrs at 37°C.
The basal expression of GFP is higher in pT7_GFP:Pveg_T7amber than the pT7_GFP control due to the leaky expression of the T7 polymerase.
In the presence of the appropriate tRNA amber suppressor, the GFP expression increases significantly, reflecting the increase in the translation of functional T7 polymerase. Therefore, the tRNA amber suppressor as well as the T7amber are fully functional with minimal leakage levels.
The T7amber was also introduced into the same strain with the T7 autoloop.
The GFP expression of this strain after an overnight culture was compared with that of a strain holding the same constructs in addition to the tRNA amber suppressor. The strains were introduced into a 96-well plate, diluted to an initial OD600 of approximately 0.1 directly in the wells. The green fluorescence and the OD600 were followed for 4hrs at 37°C.
The difference in the GFP expression is proof of a functional T7amber triggering the T7 autoloop when the appropriate tRNA amber suppressor is expressed. All final constructs for the tRNA system are functional in E.coli with behavioral profiles consistant with our expectations.
 "
"