Team:Paris Bettencourt/Experiments/YFP TetR diffusion
From 2011.igem.org
| (42 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
<html> | <html> | ||
| - | <h1> | + | <h1>Building and characterizing the YFP concentrator design</h1> |
| - | <p> | + | <p>This system is an improvement of the original experiment with the GFP. To see the fluorescence better, we decided to concentrate the YFP fluorescent molecules fused to TetR on the TetO array to make them more visible in the cell. We have been kindly given the plasmids containing the YFP:TetR construct and the TetO array by D. Lane. We biobricked the YFP:TetR construct and the TetO array so that they can be used by the synthetic biology community. The results are presented here in detail.</p> |
| + | <h2>Abstract</h2> | ||
| + | <div style="margin-left:50px; margin-right:50px; padding: 5px; border:2px solid black;"><b><p>Results for the YFP concentrator: | ||
| + | <ul> | ||
| + | <li>We successfully BioBricked both the YFP:TetR (<a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K606025">BBa_K606025</a>) and the TetO Array (<a href="http://partsregistry.org/Part:BBa_K606026">BBa_K606026</a>) constructs and sent them to the registry</li> | ||
| + | <li>We integrated our construct both in <i>E.coli</i> and <i>B.subtilis</i></li> | ||
| + | <li>We characterized the YFP:TetR fusion protein both in <i>E.coli</i> and <i>B.subtilis</i></li> | ||
| + | <li>We characterized the TetO array in <i>E.coli</i></li> | ||
| + | <li>We've done <i>E.coli</i> to <i>B.subtilis</i> diffusion experiments (with negative results)</li> | ||
| + | </ul></p></b></div> | ||
| + | <br/> | ||
<h2>Design overview</h2> | <h2>Design overview</h2> | ||
| - | <center><img src="https://static.igem.org/mediawiki/2011/9/93/YFP_concentration_principle.jpg"> | + | <center><img src="https://static.igem.org/mediawiki/2011/9/93/YFP_concentration_principle.jpg"></center> |
| - | <p> | + | <center><p>YFP:TetR/TetO array system</p></center> |
| - | <p> More information on the design | + | <p>More information on the design <a href="https://2011.igem.org/Team:Paris_Bettencourt/GFPLac_diffusion ">here</a>.</p> |
<h2>Parts and biobrick system construction</h2> | <h2>Parts and biobrick system construction</h2> | ||
| Line 21: | Line 31: | ||
<h3>TetO array construction</h3> | <h3>TetO array construction</h3> | ||
| - | <center><img src="https://static.igem.org/mediawiki/2011/a/a0/TetOarray2.jpg"> | + | <center><img src="https://static.igem.org/mediawiki/2011/a/a0/TetOarray2.jpg"></center> |
| - | <p>Cloning plan of TetO array construction</ | + | <center><p>Cloning plan of TetO array construction</p></center> |
| - | <h2> | + | <h2>Characterisation of YFP:TetR fusion protein and TetO array</h2> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <h3>Testing the YFP:tetR and | + | <h3>Testing the YFP:tetR and TetO array strains from D. Lane</h3> |
| - | In the article <a href="https://2011.igem.org/Team:Paris_Bettencourt/Experiments/YFP_TetR_diffusion#references">[1]</a>, <i>E. coli</i> strains are growing at 20°C to avoid protein | + | <p>In the article describing the YFP:TetR fusion protein<a href="https://2011.igem.org/Team:Paris_Bettencourt/Experiments/YFP_TetR_diffusion#references">[1]</a>, <i>E. coli</i> strains are growing at 20°C to avoid protein aggregation. But nanotubes between <i>B. subtilis</i> have only been observed at 37°C. |
| - | We | + | We tested different possibilities: at 37°C or 30°C and different concentrations of arabinose (0% - 0,1% -0,2%) to deal with protein aggregation.</p> |
</html> | </html> | ||
<center> | <center> | ||
{| border="1" class="wikitable" style="text-align: center;" align="center" | {| border="1" class="wikitable" style="text-align: center;" align="center" | ||
| - | |+ | + | |+YFP:TetR / TetO array : 37°C |
|- | |- | ||
| - | |[[File:TetR0_YFP02.jpg|350px|thumb|center| | + | |[[File:TetR0_YFP02.jpg|350px|thumb|center|YFP:TetR / TetO array induced with no arabinose in E. Coli from Dave Lane plasmids.]] |
| - | |[[File:TetR02_YFP03.jpg|350px|thumb|center| | + | |[[File:TetR02_YFP03.jpg|350px|thumb|center|YFP:TetR / TetO array induced with 0,2% arabinose in E. Coli from Dave Lane plasmids.]] |
|} | |} | ||
{| border="1" class="wikitable" style="text-align: center;" | {| border="1" class="wikitable" style="text-align: center;" | ||
| - | |+ | + | |+YFP:TetR / TetO array : 30°C |
|- | |- | ||
| - | |[[File:TetRTetO0_YFP01.jpg|350px|thumb|center| | + | |[[File:TetRTetO0_YFP01.jpg|350px|thumb|center|YFP:TetR / TetO array induced with no arabinose in E. Coli from Dave Lane plasmids.]] |
| - | |[[File:TetRTetO02_YFP01.jpg|350px|thumb|center| | + | |[[File:TetRTetO02_YFP01.jpg|350px|thumb|center|YFP:TetR / TetO array induced with 0,2% arabinose in E. Coli from Dave Lane plasmids.]] |
|} | |} | ||
</center> | </center> | ||
| Line 54: | Line 58: | ||
<html> | <html> | ||
| - | <p>With | + | <p>With both YFP:TetR and TetO array plasmids, we can see few spots of fluorescence with 0,2% arabinose. Because there are in the cell extremity, we can suppose that it is concentrated fluorescence in TetO arrays. Nevertheless the protein aggregation is obvious when there is only YFP:tetR at 30°C and 37°C in <i>E. coli</i>.</p> |
| - | <p>More pictures and information on the notebook | + | <p>More pictures and information on the notebook <a href="https://2011.igem.org/Team:Paris_Liliane_Bettencourt/Notebook/2011/08/03/#Kevin">here</a>.</p> |
| - | <h3>Characterization: Biobricked TetO Array | + | <h3>Characterization: Biobricked TetO Array</h3> |
| - | <h4>Microscopy of double | + | <h4>Microscopy of double transformed pFX234 / Biobricked TetO Array in <i>E. Coli</i></h4> |
| - | + | ||
| - | |||
| + | <p>To characterize this part properly, we took pictures of different strains containing TetO alone; YFP:TetR; or both TetO and YFP:TetR. In each case we made a control where the promoter was not induced with arabinose in <i>E. coli</i> (double transformated with pFX234 and TetO Array).</p> | ||
| + | </html> | ||
<center> | <center> | ||
{| border="1" class="wikitable" style="text-align: center;" align="center" | {| border="1" class="wikitable" style="text-align: center;" align="center" | ||
| - | |+ | + | |+ Biobricked TetO array only: 37°C |
|- | |- | ||
| - | |[[File:teto_minus_Fluo20.jpg|350px|thumb|center| | + | |[[File:teto_minus_Fluo20.jpg|350px|thumb|center|TetO array induced with no arabinose on E. Coli .]] |
| - | |[[File:teto_arab_Fluo20.jpg|350px|thumb|center| | + | |[[File:teto_arab_Fluo20.jpg|350px|thumb|center|TetO array induced with 0,2% arabinose on E. Coli .]] |
|} | |} | ||
{| border="1" class="wikitable" style="text-align: center;" | {| border="1" class="wikitable" style="text-align: center;" | ||
| - | |+ | + | |+ YFP:TetR only: 37°C |
|- | |- | ||
| - | |[[File:yfp_tetr_minus_fluo20.jpg|350px|thumb|center| | + | |[[File:yfp_tetr_minus_fluo20.jpg|350px|thumb|center|YFP:TetR induced with no arabinose on E. Coli .]] |
| - | |[[File:yfp_tetr_Arab_fluo20.jpg|350px|thumb|center| | + | |[[File:yfp_tetr_Arab_fluo20.jpg|350px|thumb|center|YFP:TetR induced with 0,2% arabinose on E. Coli .]] |
|} | |} | ||
{| border="1" class="wikitable" style="text-align: center;" | {| border="1" class="wikitable" style="text-align: center;" | ||
| - | |+ | + | |+YFP:TetR + Biobricked TetO array: 37°C |
|- | |- | ||
| - | |[[File:yfp_tetr_teto_minus_fluo20_2s.jpg|350px|thumb|center| | + | |[[File:yfp_tetr_teto_minus_fluo20_2s.jpg|350px|thumb|center|YFP:TetR-TetO, full construct induced with no arabinose on E. Coli .]] |
| - | |[[File:yfp_tetr_teto_Arab_fluo20_2-1.jpg|350px|thumb|center| | + | |[[File:yfp_tetr_teto_Arab_fluo20_2-1.jpg|350px|thumb|center|YFP:TetR-TetO, full construct induced with 0,2% arabinose on E. Coli .]] |
|} | |} | ||
{| border="1" class="wikitable" style="text-align: center;" | {| border="1" class="wikitable" style="text-align: center;" | ||
| - | |+ | + | |+YFP:TetR and YFP:TetR + Biobricked TetO array: 37°C - Zoom and comparison |
|- | |- | ||
| - | |[[File:yfp_tetr_zoom.jpg|350px|thumb|center| | + | |[[File:yfp_tetr_zoom.jpg|350px|thumb|center|YFP:TetR induced with 0,2% arabinose on E. Coli .]] |
| - | |[[File:yfp_tetr_teto_zoom.jpg|350px|thumb|center| | + | |[[File:yfp_tetr_teto_zoom.jpg|350px|thumb|center|YFP:TetR-TetO, full construct induced with 0,2% arabinose on E. Coli .]] |
|} | |} | ||
</center> | </center> | ||
| - | The pictures of TetO show no YFP | + | The pictures of TetO alone show no YFP fluorescence, which was expected because there is no YFP sequence in these plasmids.<br> |
| - | The TetR-YFP construct which | + | The TetR-YFP construct which (emitter part) occasionally shows gross aggregated YFP.<br> |
| - | After observing the | + | After observing the cells carrying both the TetO and YFP:TetR constructs, we can obviously distinguish glowing dots in some cells. They reflect the behavior we expected. Indeed, appearance of dots (red arrow) shows that the receiver (TetO array) actually links tightly to the YFP:TetR fusion protein! |
<h4>Microscopy of ibpA mCherry double transformated in <i>E. Coli</i></h4> | <h4>Microscopy of ibpA mCherry double transformated in <i>E. Coli</i></h4> | ||
| - | We transformated ibpA mCherry cells (expressing a mcherry in a agregation chaperon protein, | + | We wanted to be able to distinguish precisely the fusion protein aggregates from the YFP:TetR biding to TetO array. |
| + | We transformated ibpA mCherry cells (expressing a mcherry in a agregation chaperon protein, courtesy of Anne-Sophie Coquel, Inserm U1001) with pFX234 YFP:TetR and biobricked TetO Array plasmids to differenciate TetO array foci from YFP:TetR aggregates. | ||
*Case of YFP:TetR over-expression by arabinose induction | *Case of YFP:TetR over-expression by arabinose induction | ||
| Line 103: | Line 108: | ||
[[File:microscopy_yfp_ibpa.jpg|center|]] | [[File:microscopy_yfp_ibpa.jpg|center|]] | ||
| - | Microscopy shows overlap for most foci and mCherry agregation but | + | Microscopy shows overlap for most foci and mCherry agregation but some foci do not exhibit red fluorescence.<br> |
| - | Hopefully we don't expect to get high | + | Hopefully we don't expect to get high concentrations of YFP:TetR in receiver cells so aggregates will not happen in them. |
*Case of YFP:TetR low expression by arabinose induction | *Case of YFP:TetR low expression by arabinose induction | ||
| Line 110: | Line 115: | ||
[[File:microscopy_yfp_ibpa2.jpg|center|]] | [[File:microscopy_yfp_ibpa2.jpg|center|]] | ||
| - | Microscopy shows that most of | + | Microscopy shows that most of aggregates are gone and we have more not-overlaping foci.<br> |
| - | + | ||
<html> | <html> | ||
| - | < | + | <p>The ibpA experiment confirms what we suspected: <em>we can easily distinguish aggregates from YFP:TetR biding to TetO spots</em>. This means the experiments with this system can be carried on easily and will give us a clear response.</p> |
| + | |||
| + | <h3>Characterization: Biobricked YFP:TetR</h3> | ||
| + | |||
| + | <h4>Microscopy of double transformed pDAG470 / Biobricked YFP:TetR in <i>E. Coli</i></h4> | ||
| + | |||
| + | <p>To characterize this part properly, we took pictures of different strains containing YFP:TetR alone and both TetO array (from pDAG479, D.Lane) and expression YFP:TetR system (BBa_K606027).</p> | ||
</html> | </html> | ||
| - | + | <center> | |
| + | {| border="1" class="wikitable" style="text-align: center;" align="center" | ||
| + | |+ Biobricked YFP:TetR only | ||
| + | |- | ||
| + | |[[File:yfptetrbb_trans.jpg|350px|thumb|center|Biobricked expression YFP:tetR system in E. coli.]] | ||
| + | |[[File:yfptetrbb_Fluo2.jpg|350px|thumb|center|Biobricked expression YFP:tetR system in E. coli.]] | ||
| + | |} | ||
| + | {| border="1" class="wikitable" style="text-align: center;" align="center" | ||
| + | |+ Biobricked YFP:TetR and TetO Array | ||
| + | |- | ||
| + | |[[File:yfptetrbb_teto_trans.jpg|350px|thumb|center|Biobricked expression YFP:tetR system and TetO Array from D. Lane in E. coli.]] | ||
| + | |[[File:yfptetrbb_teto__Fluo2.jpg|350px|thumb|center|Biobricked expression YFP:tetR system and TetO Array from D. Lane in E. coli.]] | ||
| + | |} | ||
| + | </center> | ||
| + | <html> | ||
| + | The TetR-YFP construct which (emitter part) occasionally shows gross aggregated YFP. <br> | ||
| + | After observing the cells carrying both the TetO and YFP:TetR constructs, we can obviously distinguish glowing dots in some cells. They reflect the behavior we expected. | ||
| + | |||
| + | <h2>Testing the nanotubes with this design</h2> | ||
| + | <table> | ||
| + | <tr> | ||
| + | <td style="width:200px; text-align:center"><a href="https://2011.igem.org/Team:Paris_Bettencourt/Experiments/YFP_TetR_diffusion_experiments"><img style="width:150px" src="https://static.igem.org/mediawiki/2011/2/2d/Yfp_diff_button_pb.png"></a> | ||
| + | </td> | ||
| + | <td><b><a href="https://2011.igem.org/Team:Paris_Bettencourt/Experiments/YFP_TetR_diffusion_experiments">The YFP concentrator</a></b> This design relies on a TetO-array which allow us to concentrate YFP-TetR fusion proteins. We were able to to a <em><i>E.coli</i> to <i>B.subtilis</i> diffusion through nanotubes experiment</em> with this design. | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | </table> | ||
<html> | <html> | ||
| - | |||
<div id="citation_box"> | <div id="citation_box"> | ||
<p id="references">References and acknowledgments</p> | <p id="references">References and acknowledgments</p> | ||
| Line 126: | Line 162: | ||
</div> | </div> | ||
<br> | <br> | ||
| - | |||
<!-- PAGE FOOTER -- ITEMS FROM COLUMN ! HAVE BEEN MOVED HERE -- RDR --> | <!-- PAGE FOOTER -- ITEMS FROM COLUMN ! HAVE BEEN MOVED HERE -- RDR --> | ||
| Line 170: | Line 205: | ||
<li id="privacy"><a href="/2011.igem.org:Privacy_policy" title="2011.igem.org:Privacy policy">Privacy policy</a></li> | <li id="privacy"><a href="/2011.igem.org:Privacy_policy" title="2011.igem.org:Privacy policy">Privacy policy</a></li> | ||
| - | <li id=" | + | <li id="disclaimer"><a href="/2011.igem.org:General_disclaimer" title="2011.igem.org:General disclaimer">Disclaimers</a></li> |
| + | </ul> | ||
| + | |||
| + | </div> <!-- close footer --> | ||
| + | </div> <!-- close footer-wrapper --> | ||
| + | </div> | ||
| + | <div id="scroll_left"><a href="https://2011.igem.org/Team:Paris_Bettencourt/GFPLac_diffusion"><img src="https://static.igem.org/mediawiki/2011/0/0a/Arrow-left-big.png" style="width:100%;"></a><a href="https://2011.igem.org/Team:Paris_Bettencourt/GFPLac_diffusion">YFP concentrator design</a></div> | ||
| + | <div id="scroll_right"><a href="https://2011.igem.org/Team:Paris_Bettencourt/Experiments/YFP_TetR_diffusion_experiments"><img src="https://static.igem.org/mediawiki/2011/e/e0/Arrow-right-big.png" style="width:100%;"></a><a href="https://2011.igem.org/Team:Paris_Bettencourt/Experiments/YFP_TetR_diffusion_experiments">Testing nanotubes with the YFP concentrator system</a></div> | ||
| + | </html> | ||
Latest revision as of 00:01, 29 October 2011

Building and characterizing the YFP concentrator design
This system is an improvement of the original experiment with the GFP. To see the fluorescence better, we decided to concentrate the YFP fluorescent molecules fused to TetR on the TetO array to make them more visible in the cell. We have been kindly given the plasmids containing the YFP:TetR construct and the TetO array by D. Lane. We biobricked the YFP:TetR construct and the TetO array so that they can be used by the synthetic biology community. The results are presented here in detail.
Abstract
Results for the YFP concentrator:
- We successfully BioBricked both the YFP:TetR (BBa_K606025) and the TetO Array (BBa_K606026) constructs and sent them to the registry
- We integrated our construct both in E.coli and B.subtilis
- We characterized the YFP:TetR fusion protein both in E.coli and B.subtilis
- We characterized the TetO array in E.coli
- We've done E.coli to B.subtilis diffusion experiments (with negative results)
Design overview

YFP:TetR/TetO array system
More information on the design here.
Parts and biobrick system construction
YFP:TetR construction

Cloning plan of YFP:TetR construction
TetO array construction

Cloning plan of TetO array construction
Characterisation of YFP:TetR fusion protein and TetO array
Testing the YFP:tetR and TetO array strains from D. Lane
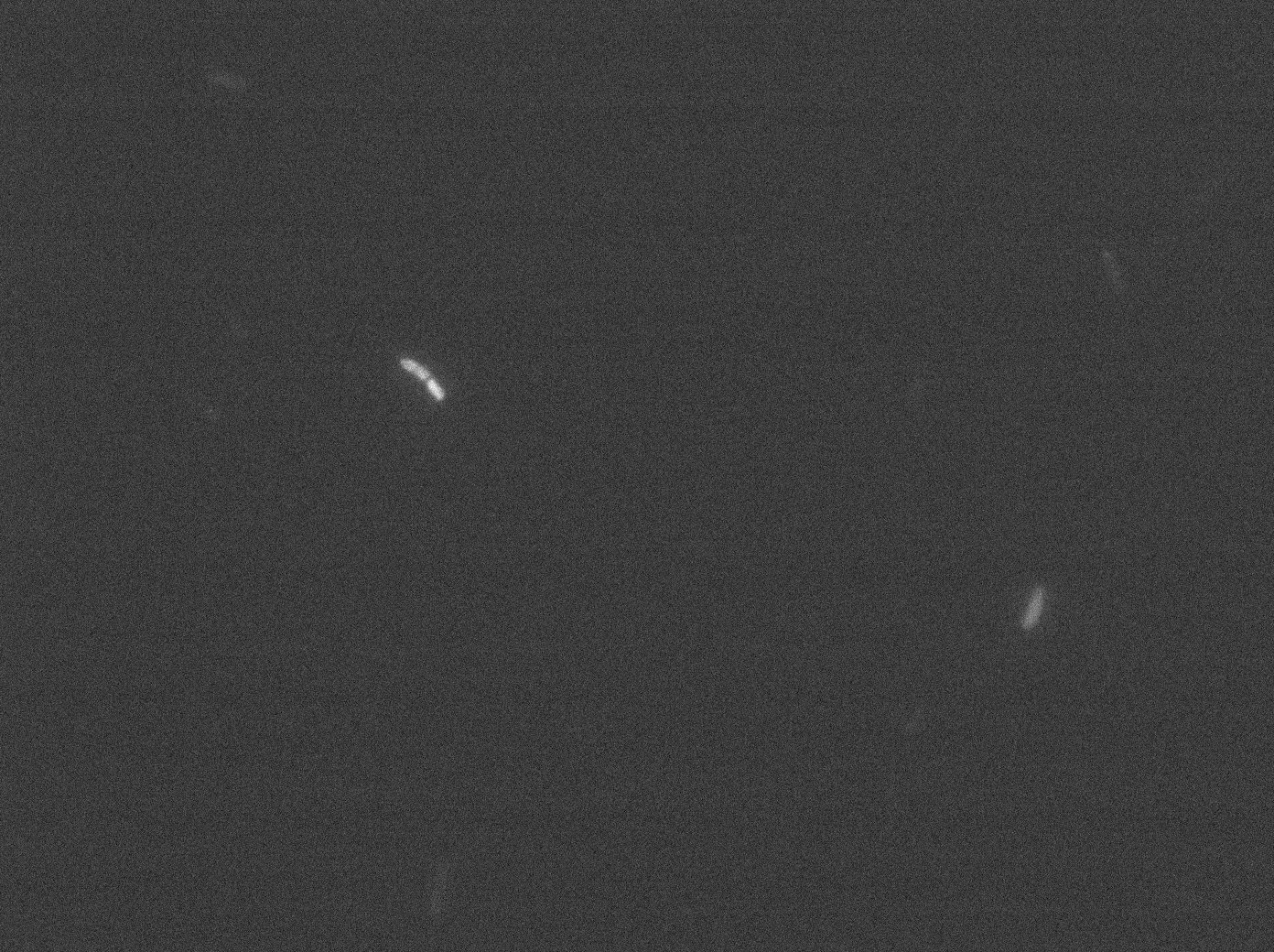
In the article describing the YFP:TetR fusion protein[1], E. coli strains are growing at 20°C to avoid protein aggregation. But nanotubes between B. subtilis have only been observed at 37°C. We tested different possibilities: at 37°C or 30°C and different concentrations of arabinose (0% - 0,1% -0,2%) to deal with protein aggregation.
With both YFP:TetR and TetO array plasmids, we can see few spots of fluorescence with 0,2% arabinose. Because there are in the cell extremity, we can suppose that it is concentrated fluorescence in TetO arrays. Nevertheless the protein aggregation is obvious when there is only YFP:tetR at 30°C and 37°C in E. coli.
More pictures and information on the notebook here.
Characterization: Biobricked TetO Array
Microscopy of double transformed pFX234 / Biobricked TetO Array in E. Coli
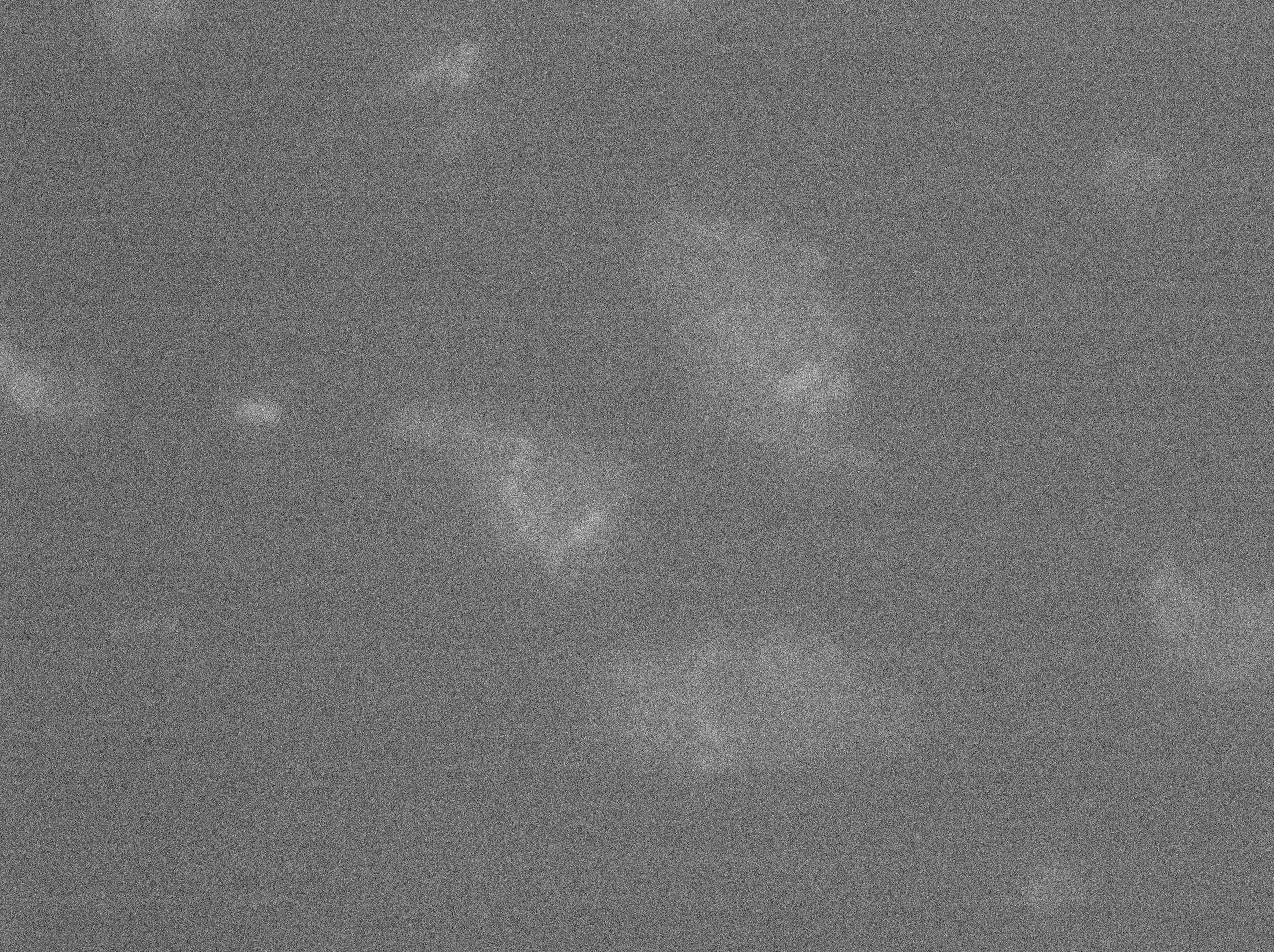
To characterize this part properly, we took pictures of different strains containing TetO alone; YFP:TetR; or both TetO and YFP:TetR. In each case we made a control where the promoter was not induced with arabinose in E. coli (double transformated with pFX234 and TetO Array).
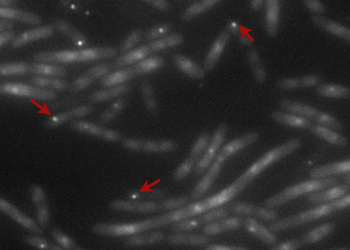
The pictures of TetO alone show no YFP fluorescence, which was expected because there is no YFP sequence in these plasmids.
The TetR-YFP construct which (emitter part) occasionally shows gross aggregated YFP.
After observing the cells carrying both the TetO and YFP:TetR constructs, we can obviously distinguish glowing dots in some cells. They reflect the behavior we expected. Indeed, appearance of dots (red arrow) shows that the receiver (TetO array) actually links tightly to the YFP:TetR fusion protein!
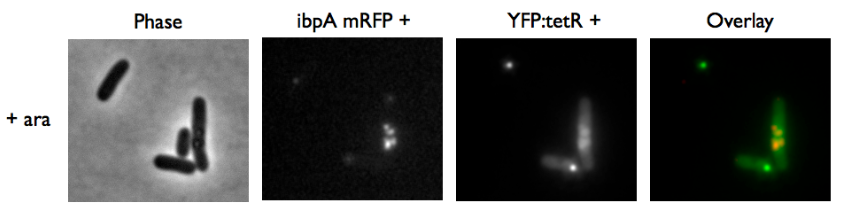
Microscopy of ibpA mCherry double transformated in E. Coli
We wanted to be able to distinguish precisely the fusion protein aggregates from the YFP:TetR biding to TetO array. We transformated ibpA mCherry cells (expressing a mcherry in a agregation chaperon protein, courtesy of Anne-Sophie Coquel, Inserm U1001) with pFX234 YFP:TetR and biobricked TetO Array plasmids to differenciate TetO array foci from YFP:TetR aggregates.
- Case of YFP:TetR over-expression by arabinose induction
Microscopy shows overlap for most foci and mCherry agregation but some foci do not exhibit red fluorescence.
Hopefully we don't expect to get high concentrations of YFP:TetR in receiver cells so aggregates will not happen in them.
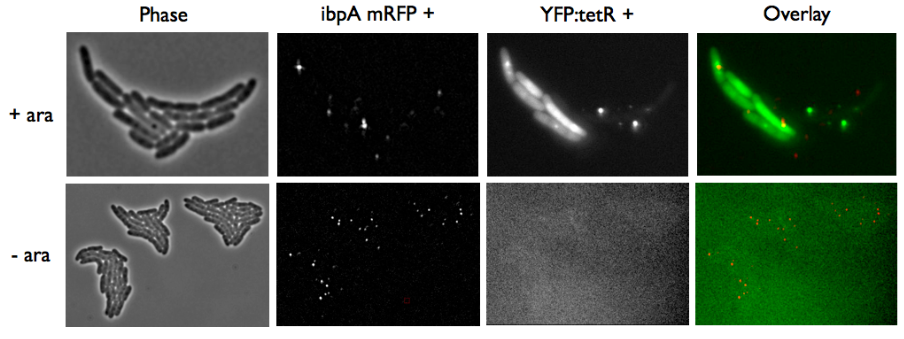
- Case of YFP:TetR low expression by arabinose induction
Microscopy shows that most of aggregates are gone and we have more not-overlaping foci.
The ibpA experiment confirms what we suspected: we can easily distinguish aggregates from YFP:TetR biding to TetO spots. This means the experiments with this system can be carried on easily and will give us a clear response.
Characterization: Biobricked YFP:TetR
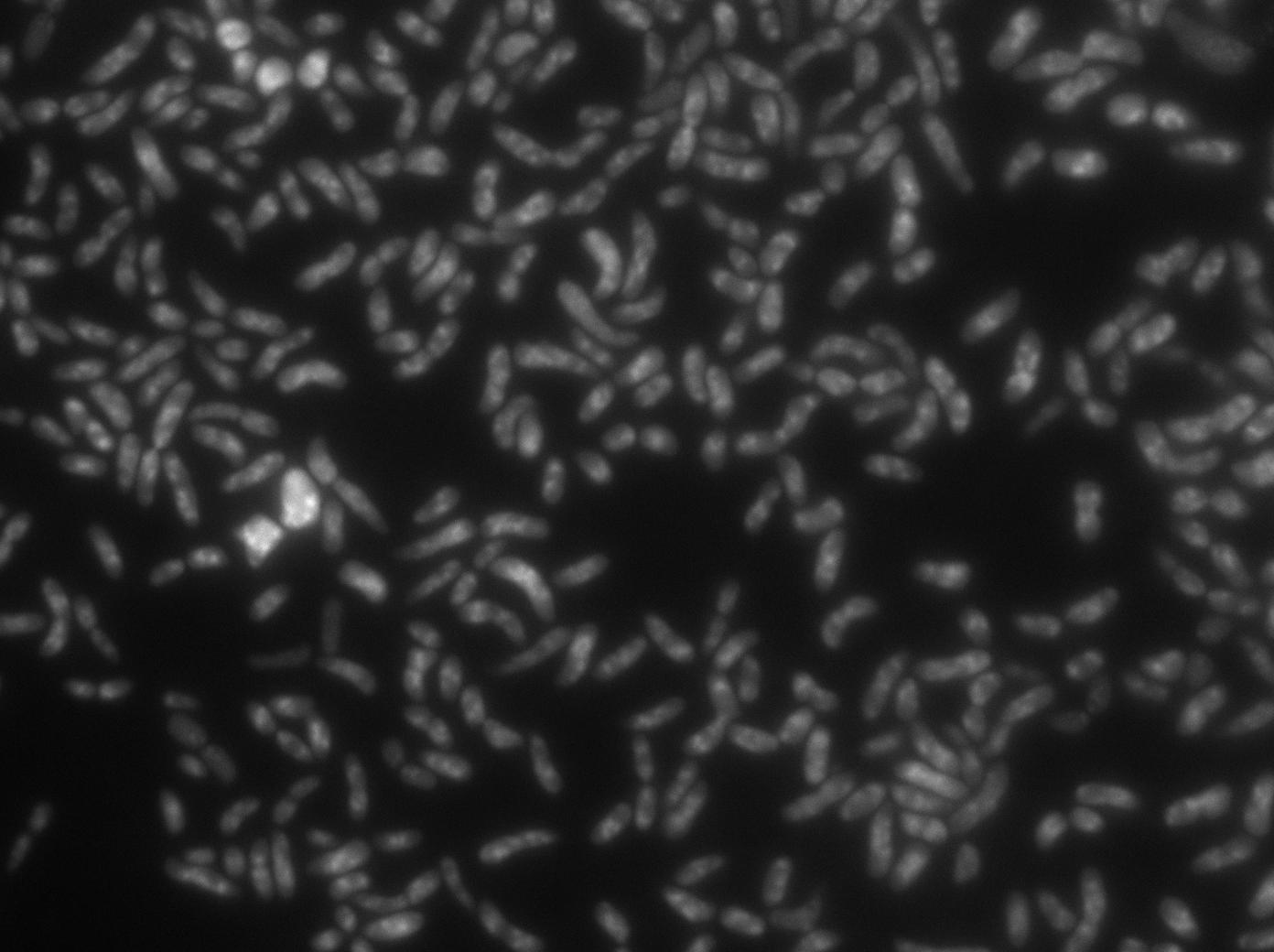
Microscopy of double transformed pDAG470 / Biobricked YFP:TetR in E. Coli
To characterize this part properly, we took pictures of different strains containing YFP:TetR alone and both TetO array (from pDAG479, D.Lane) and expression YFP:TetR system (BBa_K606027).
The TetR-YFP construct which (emitter part) occasionally shows gross aggregated YFP.
After observing the cells carrying both the TetO and YFP:TetR constructs, we can obviously distinguish glowing dots in some cells. They reflect the behavior we expected.
Testing the nanotubes with this design

|
The YFP concentrator This design relies on a TetO-array which allow us to concentrate YFP-TetR fusion proteins. We were able to to a E.coli to B.subtilis diffusion through nanotubes experiment with this design. |
References and acknowledgments
- Kinetics of plasmid segregation in Escherichia coli, Scott Gordon, Jerôme Rech, David Lane and Andrew Wright, Molecular Biology, available here Thanks to David Lane, Andrew Wright and François-Xavier Barre for information and great help they gave to us
 "
"