Team:Paris Bettencourt/Experiments/YFP TetR diffusion
From 2011.igem.org
| Line 54: | Line 54: | ||
</html> | </html> | ||
Start with the bad news : <br> | Start with the bad news : <br> | ||
| - | We have a lot of trouble to biobrick the YFP:tetR so it will be done if we successed to go to the World Jamboree. <br> | + | We have a lot of trouble to biobrick the YFP:tetR so it will be done if we successed to go to the World Jamboree. <br><br> |
But the good news : We success to biobrick the TetO array and the next step is to characterize it. The plan is to do microscopy in ''E. coli'' double transformated with YFP:tetR WT and tetO array BB, ''E. coli'' with YFP:tetR WT only (already have from D. Lane), ''E. coli'' with tetO array BB only (already have from D. Lane), Subtilis with tetO array (in pHM3). | But the good news : We success to biobrick the TetO array and the next step is to characterize it. The plan is to do microscopy in ''E. coli'' double transformated with YFP:tetR WT and tetO array BB, ''E. coli'' with YFP:tetR WT only (already have from D. Lane), ''E. coli'' with tetO array BB only (already have from D. Lane), Subtilis with tetO array (in pHM3). | ||
Revision as of 15:28, 14 September 2011

Experiments of the YFP concentration design
The planning of the experiments is the following : first we have tested the strains from D. Lane containing YFP:tetR and tetO array. Then we constructed/biobricked the YFP:tetR and tetO array system. To finish with the microscopy step and results of this proof of concept between B. subtilis and B. subtilis / E. coli.
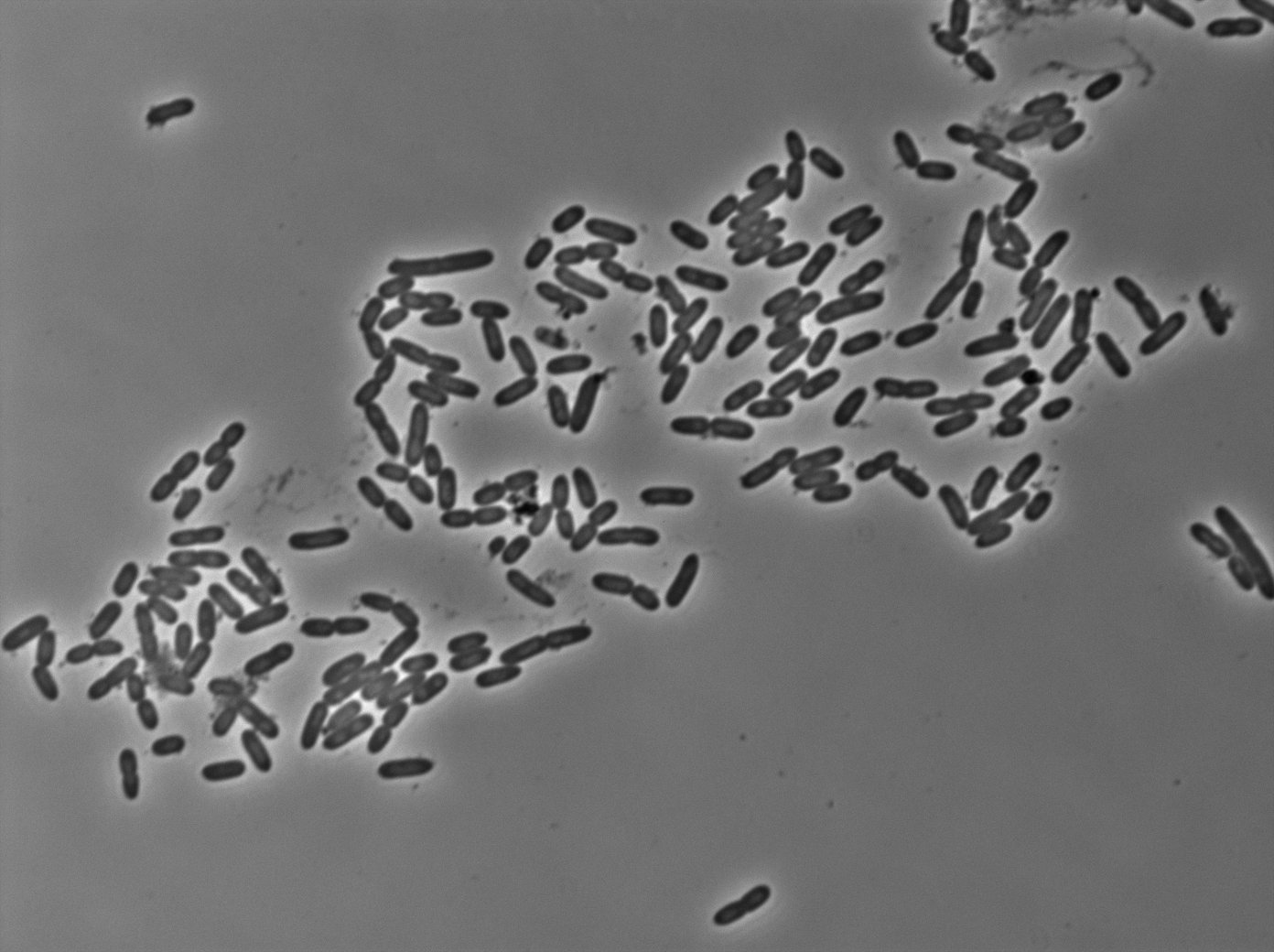
Testing the YFP:tetR strains from D. Lane
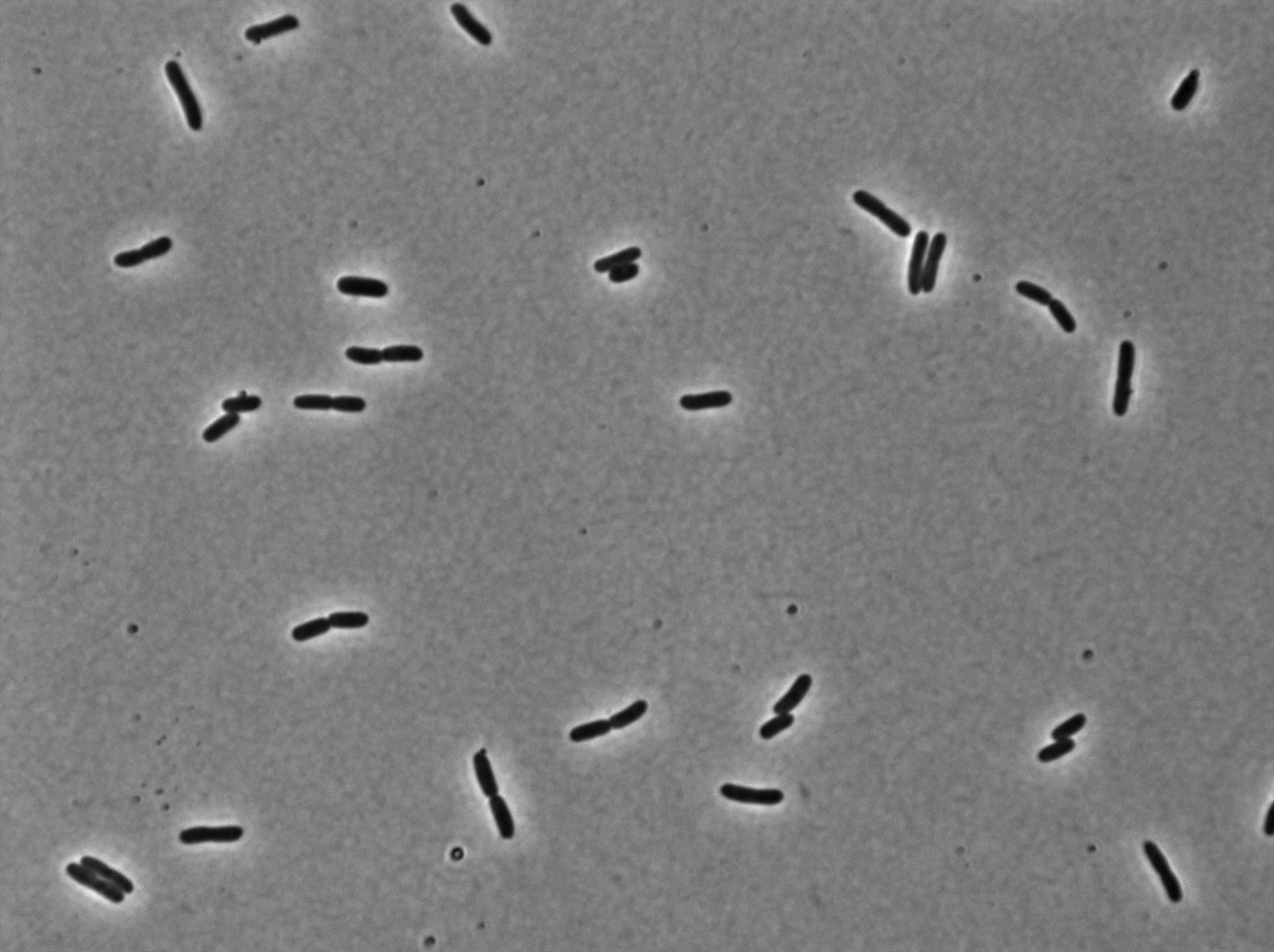
In the article [1], E. coli strains are growing at 20°C to avoid protein agregation but the problem is that nanotube between B. subtilis has been only proved to exist at 37°C. We test different possibilities : at 37°C or 30°C and concentration of arabinose (0% - 0,1% -0,2%) to deal with protein agregation.
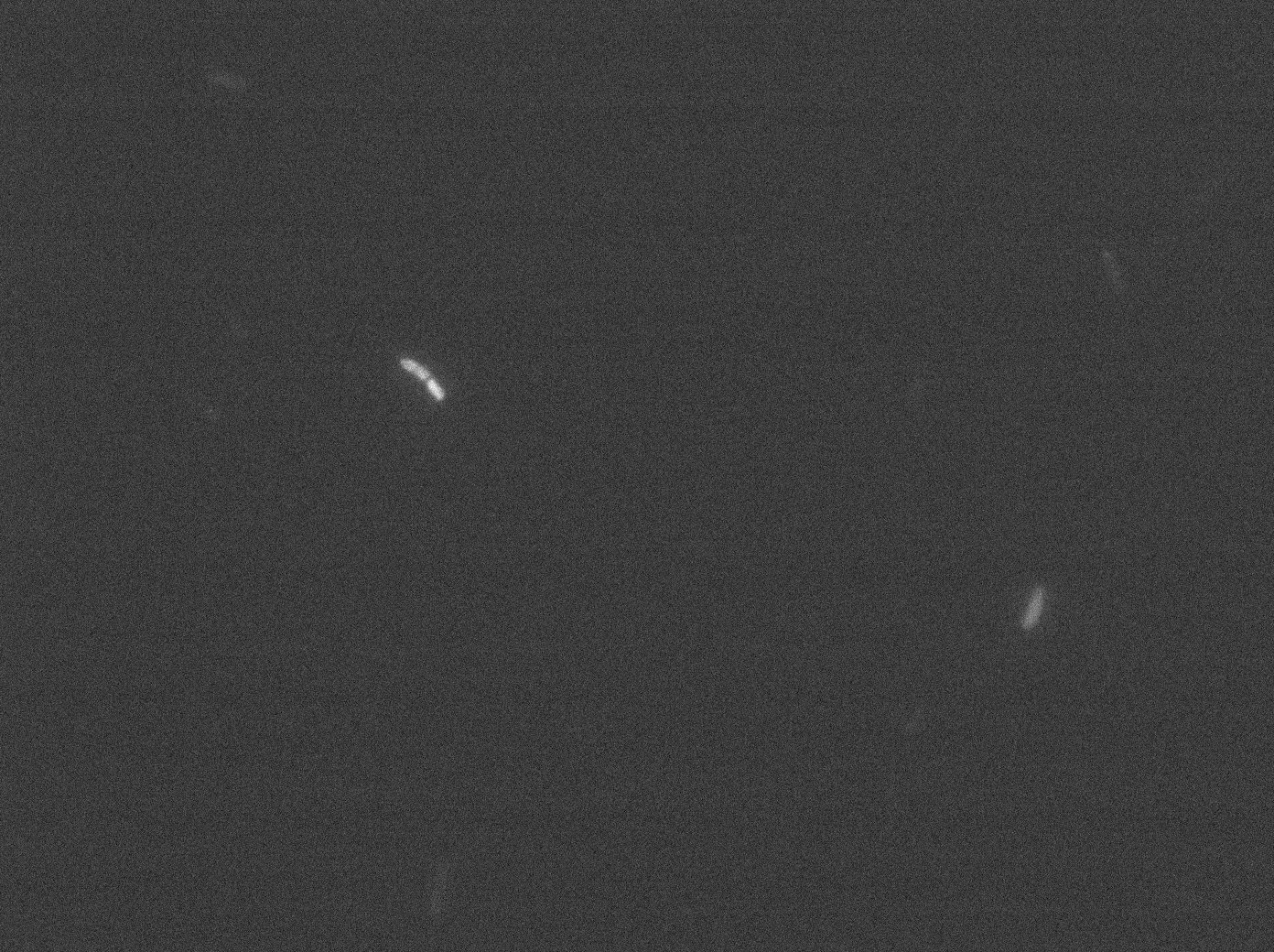
With the combinaison of YFP:tetR and TetO array plasmids, we can see few loci of fluorescence with 0,2% arabinose and because there are in the cell extremity we can suppose that it is concentrated fluorescence in tetO array. Nevertheless the protein agregation is very effective when there is only YFP:tetR at 30°C and 37°C in E. coli.
More pictures and information on the notebook [2].
Biobricked system construction
YFP:tetR construction

Fig1: Cloning plan of YFP:tetR construction
TetO array construction

Fig2: Cloning plan of TetO array construction
Results and microscopy of the proof of concept
Start with the bad news :We have a lot of trouble to biobrick the YFP:tetR so it will be done if we successed to go to the World Jamboree.
But the good news : We success to biobrick the TetO array and the next step is to characterize it. The plan is to do microscopy in E. coli double transformated with YFP:tetR WT and tetO array BB, E. coli with YFP:tetR WT only (already have from D. Lane), E. coli with tetO array BB only (already have from D. Lane), Subtilis with tetO array (in pHM3).
 "
"