Team:Paris Bettencourt/Experiments/ComS diffusion
From 2011.igem.org
| Line 12: | Line 12: | ||
<li>We successfully knocked-out CodY gene in <i>B.subtilis</i></li> | <li>We successfully knocked-out CodY gene in <i>B.subtilis</i></li> | ||
<li>We characterized the ∆CodY <i>B.subtilis</i> strain</li> | <li>We characterized the ∆CodY <i>B.subtilis</i> strain</li> | ||
| + | <li>We saw that the system does not behave as our <a href="https://2011.igem.org/Team:Paris_Bettencourt/Modeling/ComS_diffusion">model</a> predicted, with a YFP-noise level lower than expected</li> | ||
</ul></p></b></div> | </ul></p></b></div> | ||
<br/> | <br/> | ||
Revision as of 00:08, 29 October 2011

ComS/ComK switch system
The ComS/ComK switch device is based on a bistable system known as the 'MeKS' module (interactions among the MecA complex, comK and comS). ComS inhibits the MecA protease and allows the ComK protein to self-amplify. But as ComK inhibits the production of ComS, the system comes back to the original state within a few hours.
Abstract
Results for the ComK/ComS switch system:
- We successfully BioBricked both the ComS gene (BBa_K606037) and the ComS gene codon optimized (BBa_K606038) constructs and sent them to the registry
- We successfully knocked-out CodY gene in B.subtilis
- We characterized the ∆CodY B.subtilis strain
- We saw that the system does not behave as our model predicted, with a YFP-noise level lower than expected
Design overview

Fig1: Schematic of the ComS design
More information on the design here.
Parts and biobrick system construction
ComS constructions
The ComS gene was synthetized by GeneArt but also insulated by PCR colony, changing the start and the stop codon, and then cloned in pSB1C3. We then cloned in front of a pVeg-SpoVG promoter.
We didn't manage to move the construct into an integration vector and also a multihost plasmid, called pHM3 from the Harald Putzer laboratory.
Creation of the ∆CodY strain with the reporter
The genomic DNA from the CodY::spc strain from Linc Sonensheim laboratory was extracted and purified. The Elowitz's reporter strains were leaded to starvation, in order to make them competent, and then incubated for 40 min with the genomic DNA. During this time lapse, the strain is randomly making recombination between the extracellular DNA and its own genome, leading, after selection, to the KO strain.
Then, the B. Subtilis are selected on Spectinomycine plates, and only the KO clones were selected. This protocol works very well and we got a huge number of recombinants.
Characterization of the ComS biobrick
The construct was cloned into our multihost vector. Electroporation into Elowitz reporter strain would permit to test the construct. Indeed, YFP expression would indicate that the ComS gene created is active and CFP expression reports that the ComK production has been triggered by overexpression of ComS.
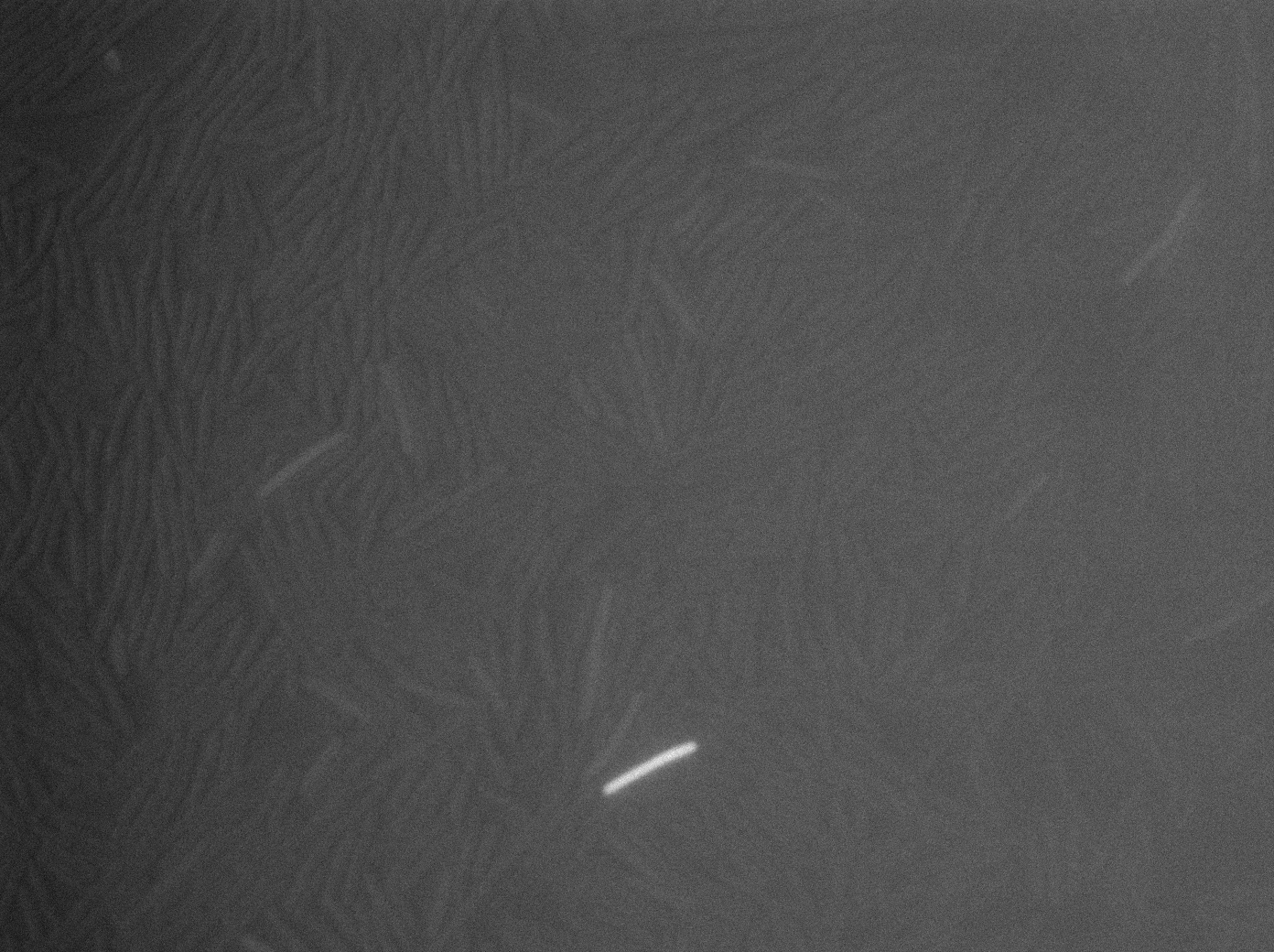
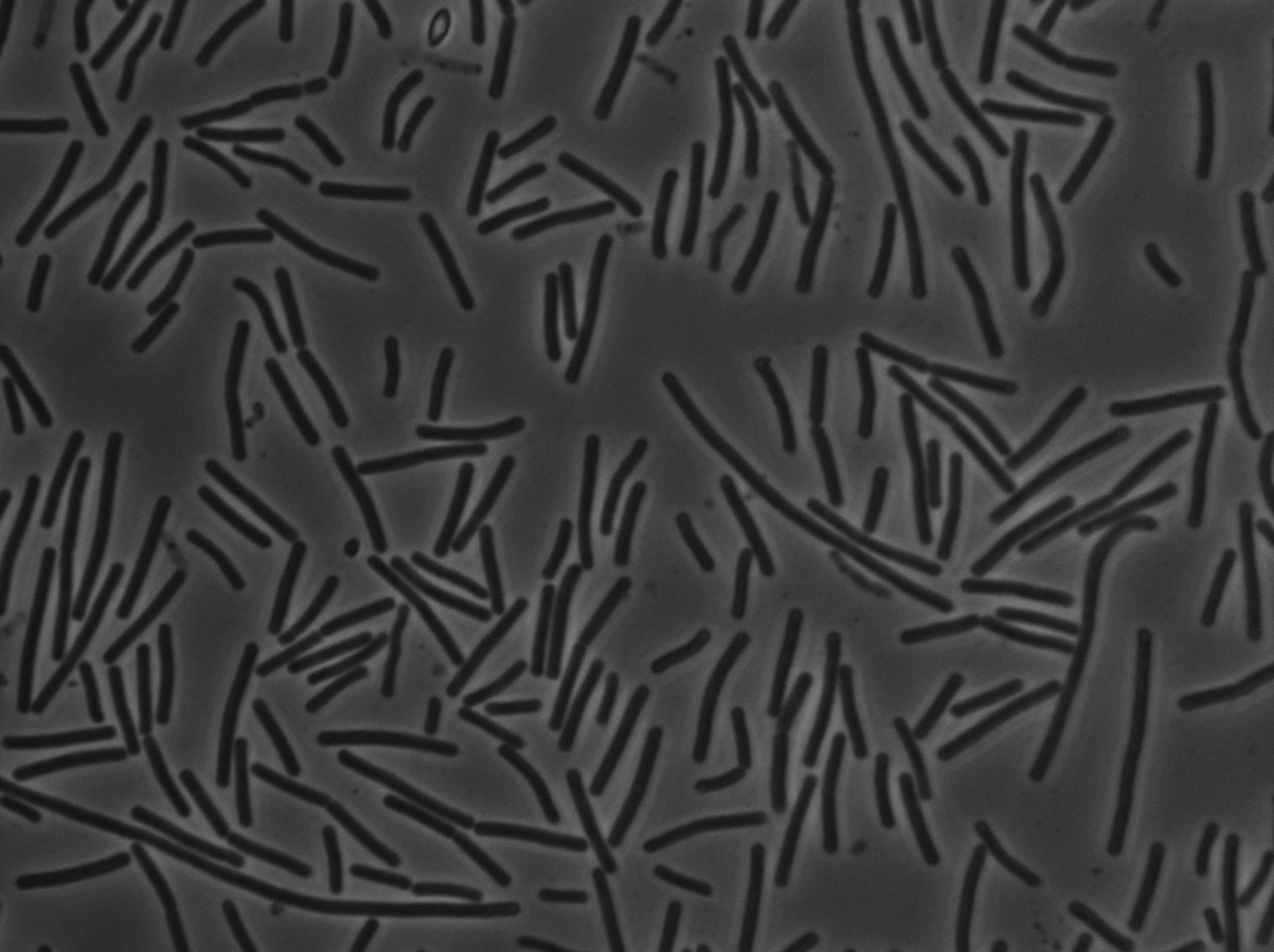
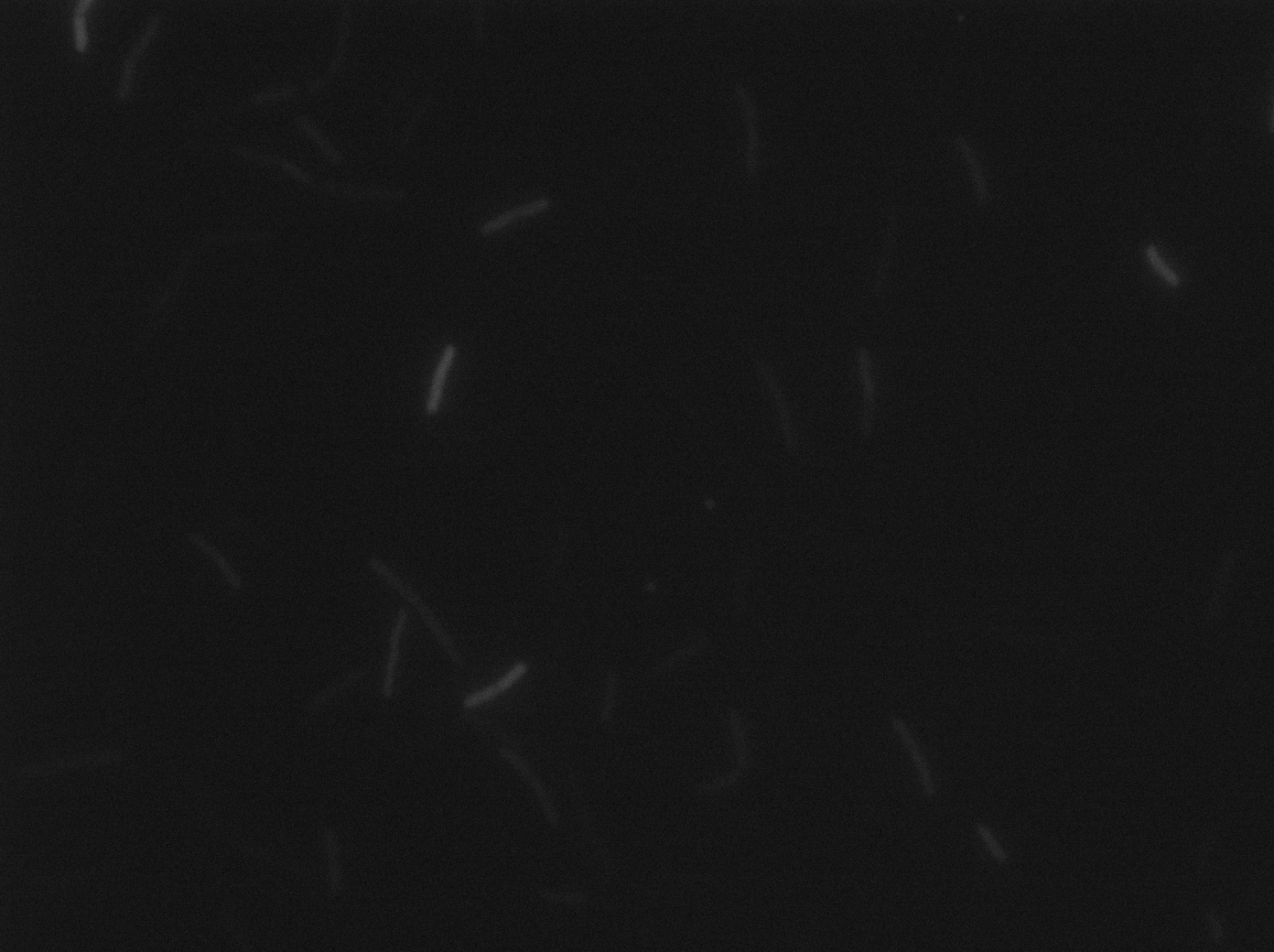
Characterization of the ∆CodY strain
We characterized ∆CodY B. subtilis strain.
These pictures show that the T7 GFP autoloop system is efficient since some cells are glowing with GFP fluorescence. Thus, we can conclude that the T7 autoloop is activated because of stochastic leakage.
Finally, we notice there is no difference between the GFP autoloop with and without terminator before the T7 promoter. It could be due to that terminator which is a B.subtilis terminator. Moreover, we know that this E.coli plasmid has 4 terminators before our construct, pretty much nullifying the effect of our extra terminator in E.coli.
 "
"