Team:NTNU Trondheim/rrnB
From 2011.igem.org
(→rrnB P1) |
(→ppGpp's effect on rrnB P1 promoter) |
||
| Line 98: | Line 98: | ||
'''[2]''' Tedin, K., A. Witte, et al. (1995). "Evaluation of the E. coli ribosomal rrnB P1 promoter and phage-derived lysis genes for the use in a biological containment system: A concept study." Journal of Biotechnology 39(2): 137-148. | '''[2]''' Tedin, K., A. Witte, et al. (1995). "Evaluation of the E. coli ribosomal rrnB P1 promoter and phage-derived lysis genes for the use in a biological containment system: A concept study." Journal of Biotechnology 39(2): 137-148. | ||
| + | |||
{{:Team:NTNU_Trondheim/NTNU_footer}} | {{:Team:NTNU_Trondheim/NTNU_footer}} | ||
Revision as of 18:16, 20 September 2011

rrnB P1
Previous studies have shown that the rrnB P1 promoter is downregulated by ppGpp[1]. That is why we decided to use this promoter as the stress sensing promoter in the stress sensor. A version of rrnB P1 already existed in the Registry of Standard Biological Parts. This version was made by the Berkeley iGEM team in 2008.
The [http://partsregistry.org/Part:BBa_K112118 BioBrick] that the Berkeley team made is in [http://openwetware.org/wiki/Template:AndersonLab:BBb_Standard BBb format]. This means that the BioBrick is flanked by a prefix and a suffix that is different from the ones that are normally used. Therefore, this BioBrick was initially not compatible with the other BioBricks we were planning to use in our construct, as all the other BioBricks we planned to use were in BBa format.
To deal with this problem, the rrnB P1 promoter was amplified using PCR. The original promoter made by the Berkeley iGEM team was used as template DNA. To make the amplified brick compatible with the rest of our construct, the [http://partsregistry.org/Help:BioBrick_Prefix_and_Suffix standard prefix and suffix] was added to our primers.
When we amplified the promoter, we amplified both the whole brick (rrnB P1) and only the assumed promoter sequence (proL). Primers for both amplifications are given below:
rrnB P1.fwd: GTTTCTTCGAATTCGCGGCCGCTTCTAGAGACGTATCCTACGCCCGTGGT
rrnB P1.rev: GTTTCTTCCTGCAGCGGCCGCTACTAGTACGCCTTCCCGCTACAGAGTCA
proL.fwd: GTTTCTTCGAATTCGCGGCCGCTTCTAGAGCCTCTTGTCAGGCCGGAATAACTCC
proL.rev: GTTTCTTCCTGCAGCGGCCGCTACTAGTAGCGGCGTGTTTGCCGTTGTT
The nucleotide sequences of both PCR products are given below:
Nucleotide sequence of rrnB P1 biobrick
Nucleotide sequence of proL biobrick
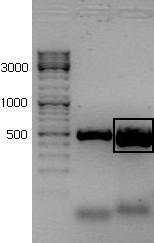
The PCR-product was run on a 1,5% agarose gel in order to verify that we had achieved the correct product and to separate it from any other unwanted products.
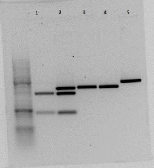
The new rrnB P1 biobrick was inserted in the psb1C3 plasmid provided by the iGem HQ. Plasmid from five colonies were digested with the enzymes BstBI and SpeI It seems like parallel 1 and 2 have the insert.
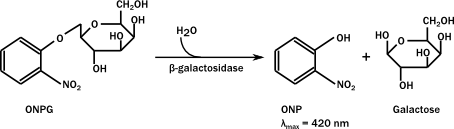
We planned to characterize the promoter using a construct consisting of rrnB P1, LacZ, pBad and relA. pBad is an inducable promoter that gets upregulated in response to the presence of arabinose. relA was set under control of the pBad promoter. When the promoter got upregulated due to arabinose, relA would be transcribed, resulting in ppGpp. To see if ppGpp really downregulates the rrnB P1 promoter, this promoter was coupled to the LacZ gene. LacZ codes for β-lactamase, which is capable of cleaving ONPG, yielding a yellow color. For the results of the caracterization, see the characterization page.
Link to parts registry: [http://partsregistry.org/Part:BBa_K639002 BBa_K639002]
ppGpp's effect on rrnB P1 promoter
Abstract
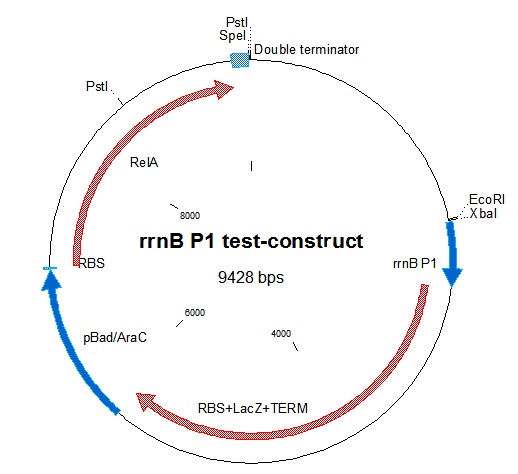
To test the down-regulating effect of ppGpp on rrnB P1, which is the basis of our project, we decided to make a construct containing ppGpp Synthase (RelA) inducible by the pBAD/AraC promoter. The construct would also contain beta-galactosidase (lacZ) which would be expressed by the rrnB P1 promoter. Thus, induction of the pBAD/AraC promoter with arabinose should give lower lacZ production, as relA is overproduced giving high ppGpp concentration. We were not able to show this effect in our experiment due to an error in the cloning procedure.
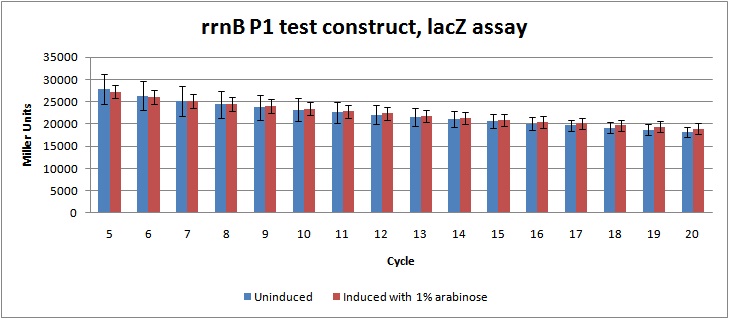
Figure 2: LacZ assay data in Miller units from rrnB P1 test-construct, cycle 5-20. Showing no significant difference between the induced cultures (which should be lower due to prduction of relA) and uninduced cultures.
Unfortunately, the data from the assay indicates no significant regulation of the rrnB P1 promoter, as there is no difference between the induced and the uninduced cultures. This might be due to problems during the genetic construction, as enzyme test cutting of the construct didn't give the expected fragments. The construct will be sequenced to figure out what went wrong.
UPDATE: It was clear from the sequencig results that the construct had errors. It consisted only of half the RelA gene and no pBad promoter.
Another problem was that it seemed like the lacZ construct had constitutive expression without a promoter. The transformants containing only the [http://partsregistry.org/Part:BBa_I732019 lacZ BioBrick] gave blue colonies on X-gal, and analysis of the BioBricks sequence showed promoter-like features ahead of the actual lacZ gene in some web-tools for promoter analysis. As the rrnB P1 promoter is quite weak in our constructs (see stress-sensor), it could be that the difference in rrnB P1 expression is over-shadowed by the constitutive expression of the lacZ BioBrick.
ONPG assay
β-galactosidase hydrolyzes ortho-Nitrophenyl-β-galactoside (ONPG) to ONP + galactoside. ONP gives a yellow color, and absorbs at 420 nm. LacZ assays are widely used to test promoter activity. Here, the assay was performed as follows;
Cells were grown over night in pre-culture, inoculated 1% in appropriate media in 3 parallells and grown for 18 hours. 5µL culture was transferred to a 96 well plate and 100 μl of Z-buffer with chloroform (Z-buffer: 0,06 M Na2HPO4 x 7H20, 0,04 M NaH2PO4 x H20, 0,1M KCl, 0,001 M MgSO4x7H2O, pH 7; Z-buffer with chloroform: Z-buffer, 1% β-mercaptoethanol, 10% chloroform)was added. 50 µL Z-buffer with SDS 1,6% was added to lyse the cells, and the plate was incubated for 10 minutes in room temperature. 50 µL 0,4 % ONPG solution in Z-buffer was added, and OD405 was measured with 1 minute intervals for 20 cycles, in 4 parallells.
Miller units were calculated as shown in [http://openwetware.org/wiki/Beta-Galactosidase_Assay_(A_better_Miller) Openwetware's A better Miller], to compensate for OD and reaction time. Figure 2 shows the Miller units from cycle 5-20.
Genetic construction
Our relA BioBrick was digested with E+S, it was ligated into E+X-digested [http://partsregistry.org/partsdb/get_part.cgi?part=BBa_B0015 BBa_B0015]. the relA+double terminator was then ligated into an RBS backbone from [http://partsregistry.org/wiki/index.php/Part:BBa_B0034 BBa_B0034]. To be able to control the expression of relA, the arabinose-inducible promoter/inducer [http://partsregistry.org/Part:BBa_K113009 BBa_K113009] containing pBAD/AraC was put in front of the construct.
rrnB P1 was joined with [http://partsregistry.org/Part:BBa_I732019 BBa_I732019] containing an RBS, lacZ coding for beta-galactosidase, and a double terminator. This was then combined with the pBAD-relA construct. A problem we encountered was that relA contains a PstI site at bp 1393. This was mostly worked around in our construction. In one step in the construction, we had to digest relA with PstI. In that case, partial digestion was used. The final construct is represented in figure 1.
[1] Magnusson, Lisa U., Farewell, Anne, Nyström, Thomas: "ppGpp: a global regulator in Escherichia Coli", 2005
[2] Tedin, K., A. Witte, et al. (1995). "Evaluation of the E. coli ribosomal rrnB P1 promoter and phage-derived lysis genes for the use in a biological containment system: A concept study." Journal of Biotechnology 39(2): 137-148.
 "
"